Track and Quantitate Exosomes with Antibodies and ELISAs
Exosomes are small membrane vesicles secreted by most cell types in vivo and in vitro. Exosomes are found in cell culture media, blood, urine, amniotic fluid, malignant ascite fluids and contain distinct subsets of microRNAs and proteins depending upon the tissue from which they are secreted. SBI’s ExoELISA kits are designed for fast and quantitative analysis of three well-characterized exosomal protein markers: CD63, CD9, CD81 or Hsp70.
The exosome antibody kits (cat#s EXOABxxx) allow for the confirmation of exosome recoveries by Western blots. SBI has also built an Exo-Check™ Exosome Antibody Array (cat# EXORAY) that allows you to to detect 8 known exosome markers and a GM130 cis-Golgi marker to monitor any cellular contamination simultaneously in your exosome isolations. The ExoELISA kits (cat# EXOEL) enable the specific quantitation of CD63, CD9 or CD81 positive exosome microvesicles. The exosome antibodies, antibody arrays and ExoELISA kits are fully compatible with exosomes isolated by SBI’s ExoQuick or ExoQuick-TC as well as ultracentrifugation methods.
Exosome antibodies for Western blot analysis
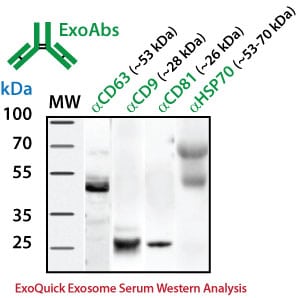
For Western blotting analysis, we recommend resuspending the exosome pellet in 1XRIPA buffer with the appropriate protease inhibitor cocktail. SBI offers individual antibodies for CD63, CD9, CD81 and Hsp70 as well as a Western blot sampler kit (Catalog# EXOAB-KIT-1) which includes four exosomal marker antibodies: CD63, CD9, CD81, HSP70 (rabbit anti-human) and also includes a goat anti-Rabbit IgG HRP conjugated secondary antibody specifically tested for use in exosomal protein analysis. The primary antibodies are used at a 1:1,000 dilution and the HRP secondary antibody at 1:20,000 dilution.
Exosome Marker Protein Analysis
|
Target
|
Catalog #
|
# Blots (5ml)
|
| CD63 | EXOAB-CD63A-1 |
5
|
| CD9 | EXOAB-CD9A-1 |
5
|
| CD81 | EXOAB-CD81A-1 |
5
|
| Hsp70 | EXOAB-Hsp70A-1 |
5
|
| All Four | EXOAB-KIT-1 |
5
|
Exo-Check™ exosome antibody arrays
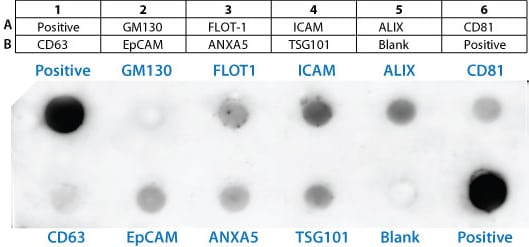
The Exo-Check antibody array has 12 pre-printed spots and features 8 antibodies for known exosome markers (CD63, CD81, ALIX, FLOT1, ICAM1, EpCam, ANXA5 and TSG101) and a GM130 cis-Golgi marker to monitor any cellular contamination in your exosome isolations. The array is compatible with ultracentrifugation and ExoQuick methods. The positive control spot is derived from human serum exosome proteins and are printed directly onto the membrane. Your exosome preparations are lysed and then incubated with the array for the pre-printed antibodies to capture their respective exosome proteins. The GM130 antibody spot on each array is included to help identify and cellular contamination in your exosome preparations. The kits come complete with a secondary detection mixture conjugated to HRP. The arrays are semi-quantitative and can be used to evaluate relative abundance of certain exosome protein markers from a set of given samples. The orientation of the Array is identified as using the upper left hand corner that is notched.
Sample data
The positive control signals should provide a bright signal and this will indicate that all of the detection reagents are working properly. The various exosome antibody spots will provide signals of varying degree, depending upon the source of the isolated exosomes. The Blank spot should not have any signal and this serves as a background control. The GM130 control is to evaluate cellular contamination (if any) in your exosome preparations. The data below are from 300 ug of exosome proteins isolated using ExoQuick-TC from human HT1080 lung sarcoma cell line media.
Information about the exosome protein targets
The table below shows each of the exosome proteins that can be detected on the array along with a link to the ExoCart protein database.
Location |
ID |
Gene Name |
Exocarta Link/Comments |
|
A1
|
Positive | Positive control for HRP detection | |
|
A2
|
GM130 | cis-Golgi matrix protein | Monitor cellular contamination in exosome preps |
|
A3
|
FLOT1 | Flotillin 1 | https://exocarta.org/gene_summary?gene_id=10211 |
|
A4
|
ICAM1 | Intercellular adhesion molecule 1 | https://exocarta.org/gene_summary?gene_id=3383 |
|
A5
|
ALIX | PDCD6IP, Programmed cell death 6 interacting protein, AIP1/DRIP4/HP95 | https://exocarta.org/gene_summary?gene_id=10015 |
|
A6
|
CD81 | IGSF8, Immunoglobulin superfamily, member 8 | https://exocarta.org/gene_summary?gene_id=93185 |
|
B1
|
CD63 | CD63, LAMP-3 | https://exocarta.org/gene_summary?gene_id=967 |
|
B2
|
EpCAM | Epithelial cell adhesion molecule | https://exocarta.org/gene_summary?gene_id=4072 |
|
B3
|
ANXA5 | Annexin A5 | https://exocarta.org/gene_summary?gene_id=308 |
|
B4
|
TSG101 | Tumor susceptibility gene 101 | https://exocarta.org/gene_summary?gene_id=7251 |
|
B5
|
BLANK | No antibody loaded to test for background | |
|
B6
|
Positive |
Quantitate Exosomes by ELISA
The ExoELISA kit is designed as a direct Enzyme-Linked ImmunoSorbent Assay (ELISA). The exosome particles and their proteins are directly immobilized onto the wells of the microtiter plate. After binding, wells are coated with a block agent to prevent non-specific binding of the detection antibody. The detection (primary) antibody is added to the wells for binding to specific antigen (e.g. CD63) protein on the exosomes.
Choose from 3 different primary antibody detection formats:
- CD63 ExoELISA, catalog# EXOEL-CD63A-1
- CD9 ExoELISA, catalog# EXOEL-CD9A-1
- CD81 ExoELISA, catalog# EXOEL-CD81A-1
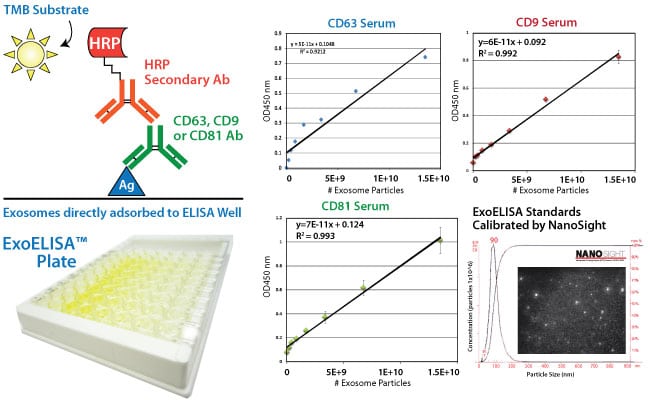
A Horseradish Peroxidase enzyme (HRP) linked secondary antibody (goat anti-rabbit) is used for signal amplification and to increase assay sensitivity. A colorimetric substrate (Extra-sensitive TMB) is used for the assay read-out. The accumulation of the colored product is proportional to the specific antigen present in each well. The results are quantitated by a microtiter plate reader at 450 nm absorbance and calibrated by the exosome standards provided in the kit. The exosome standards are provided in number of exosome particles as determined by NanoSight Nanoparticle Tracking Analysis. The ExoELISA kits are compatible with exosomes isolated by ExoQuick, ExoQuick-TC or by ultracentrifugation techniques.
Online Resources
- Exosome Antibody and ELISA Kits User Manual (PDF)
- Exocarta online database of exosome proteins and RNAs
Product Citations
- Ramteke A, Ting H, Agarwal C, Mateen S, Somasagara R, Hussain A, Graner M, Frederick B, Agarwal R, Deep G. Exosomes secreted under hypoxia enhance invasiveness and stemness of prostate cancer cells by targeting adherens junction molecules. Mol Carcinog. 2013 Dec 17. doi: 10.1002/mc.22124.
- Dubovsky JA, Chappell DL, Harrington BK, Agrawal K, Andritsos LA, Flynn JM, Jones JA, Paulaitis ME, Bolon B, Johnson AJ, Byrd JC, Muthusamy N. Lymphocyte cytosolic protein 1 is a chronic lymphocytic leukemia membrane-associated antigen critical to niche homing. Blood. 2013 Nov 7;122(19):3308-16.
- Masato Mitsuhashi, Dennis D. Taub, Dimitrios Kapogiannis, Erez Eitan, Linda Zukley, Mark P. Mattson, Luigi Ferrucci, Janice B. Schwartz, and Edward J. Goetzl. Aging enhances release of exosomal cytokine mRNAs by Aβ1-42-stimulated macrophages. FASEB J. 2013; 27:5141-5150.
- Sohel MMH, Hoelker M, Noferesti SS, Salilew-Wondim D, Tholen E, et al. Exosomal and Non-Exosomal Transport of Extra-Cellular microRNAs in Follicular Fluid: Implications for Bovine Oocyte Developmental Competence. PLoS ONE 8(11): e78505.
- Alhasan, A. H., Patel, P. C., Choi, C. H. J. and Mirkin, C. A. Exosome Encased Spherical Nucleic Acid Gold Nanoparticle Conjugates as Potent MicroRNA Regulation Agents. Small. doi: 10.1002/smll.201302143. (PDF) »
- Lee HK, Finniss S, Cazacu S, Bucris E, Ziv-Av A, Xiang C, Bobbitt K, Rempel SA, Hasselbach L, Mikkelsen T, Slavin S, Brodie C. Mesenchymal stem cells deliver synthetic microRNA mimics to glioma cells and glioma stem cells and inhibit their cell migration and self-renewal. Oncotarget. 2013 Feb;4(2):346-61.
- Ariza ME, Rivailler P, Glaser R, Chen M, Williams MV. Epstein-Barr Virus Encoded dUTPase Containing Exosomes Modulate Innate and Adaptive Immune Responses in Human Dendritic Cells and Peripheral Blood Mononuclear Cells. PLoS ONE 8(7): e69827.
- Chugh PE, Sin S-H, Ozgur S, Henry DH, Menezes P, et al. Systemically Circulating Viral and Tumor-Derived MicroRNAs in KSHV-Associated Malignancies. PLoS Pathog 9(7): e1003484. doi:10.1371/journal.ppat.1003484.
- Bashratyan R, Sheng H, Regn D, Rahman MJ, Dai YD. Insulinoma-released exosomes activate autoreactive marginal zone-like B cells that expand endogenously in prediabetic NOD mice. Eur J Immunol. 2013 Jul 2. doi: 10.1002/eji.201343376.
- Epple LM, Griffiths SG, Dechkovskaia AM, Dusto NL, White J, Ouellette RJ, Anchordoquy TJ, Bemis LT, Graner MW. Medulloblastoma Exosome Proteomics Yield Functional Roles for Extracellular Vesicles. (2012) PLoS ONE 7(7): e42064. doi:10.1371/journal.pone.0042064. (PDF) »
- Singh PP, Smith VL, Karakousis PC, Schorey JS. Exosomes Isolated from Mycobacteria-Infected Mice or Cultured Macrophages Can Recruit and Activate Immune Cells In Vitro and In Vivo. J Immunol. 2012 Jun 20. [Epub ahead of print].
- Alvarez ML, Khosroheidari M, Kanchi Ravi R, Distefano JK. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012 Jul 11. doi: 10.1038/ki.2012.256. (PDF) »
Technical References
- Mathivanan, S. and Simpson, R.J. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics. 2009.21, 4997-5000.
- Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009. 8, 581-93.
- Adachi T, Nakanishi M, Otsuka Y, Nishimura K, Hirokawa G, Goto Y, Nonogi H, Iwai N. Plasma microRNA 499 as a biomarker of acute myocardial infarction. Clin Chem. 2010 Jul;56(7):1183-5.
- De Smaele E, Ferretti E, Gulino A. MicroRNAs as biomarkers for CNS cancer and other disorders. Brain Res. 2010 Jun 18;1338:100-11.
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008 Jul 29;105(30):10513-8.
- Laterza OF, Lim L, Garrett-Engele PW, Vlasakova K, Muniappa N, Tanaka WK, Johnson JM, Sina JF, Fare TL, Sistare FD, Glaab WE. Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem. 2009 Nov;55(11):1977-83.
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007 Jun;9(6):654-9.
- Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Wordinger T, Middeldorp JM. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA; 2010 Apr 6; 107(14):6328-33.
- Michael A, Bajracharya SD, Yuen PS, Zhou H, Star RA, Illei GG, Alevizos I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis; 2010 Jan; 16(1):34-8.
- Luo SS, Ishibashi O, Ishikawa G, Ishikawa T, Katayama A, Mishima T, Takizawa T, Shigihara T, Goto T, Izumi A, Ohkuchi A, Matsubara S, Takeshita T, Takizawa T. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol Reprod; 2009 Oct; 81(4):717-29.
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol; 2008 Jul; 110(1):13-21.
- Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009 Jun;6(3):267-83. Review.