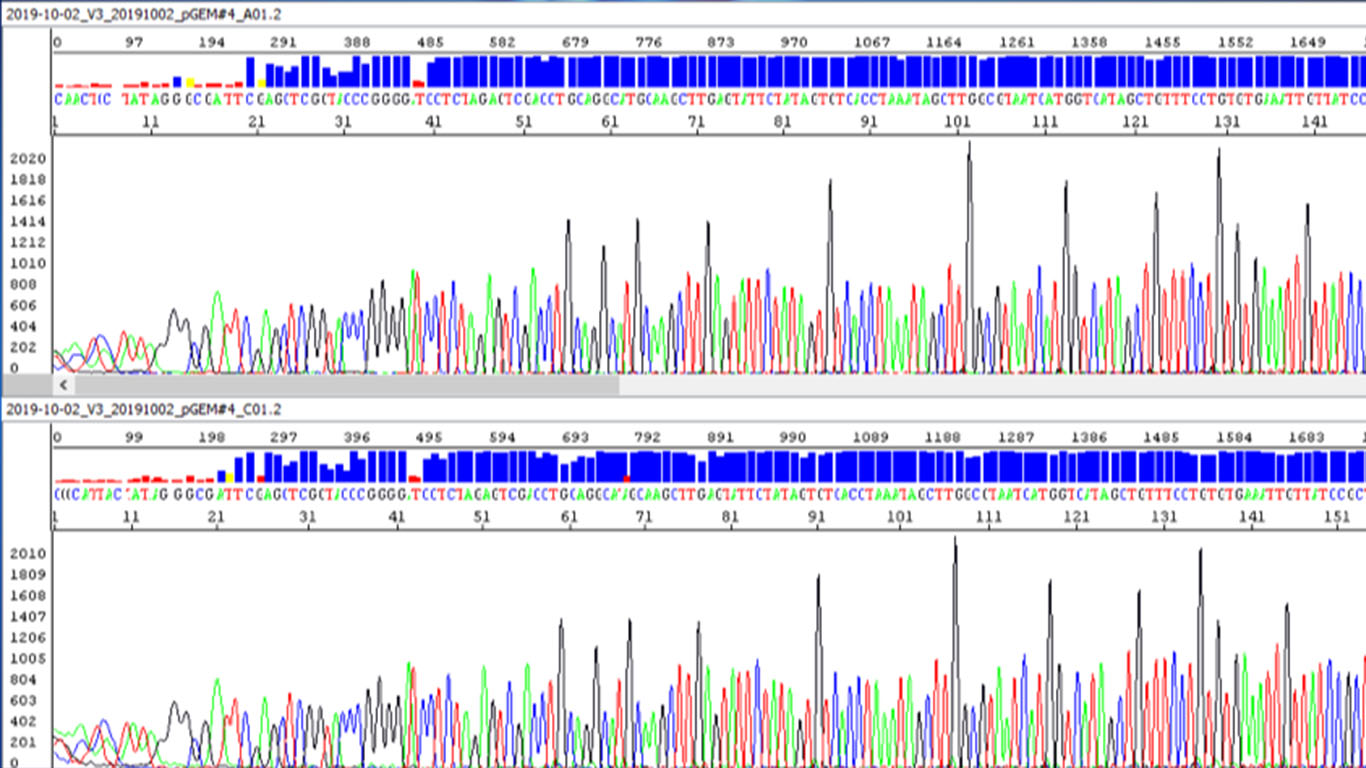
The sample (DNA template) purity and quantity are the most important factors affecting the success of sequencing results. As for purity, the template DNA should not be contaminated with proteins, polysaccharides, RNAs, or genomic DNA. As for the quantity, a proper DNA quantification method other than a mere spectrometer for 260 nm and 280 nm measurement should be used to measure the concentration of the template DNA. The right amount of template based on the length of the template should be used in the sequencing reaction. Most common Sanger sequencing templates could be a plasmid, PCR products or DNA made from Rolling Cycle Amplification (RCA).
Plasmid used as template
Make sure plasmid of high quality is prepared for optimal performance. Plasmids that can be used as PCR products or digested with restriction enzymes may still not be good enough as Sanger sequencing templates. During plasmid preparation, it is critical to wash the purification columns thoroughly and elute DNA after removing traces of ethanol in the column.
PCR products used as template
PCR products are more favored as sequencing templates, but they should be of high purity and uniformity. Make sure high quality of single and specific PCR products are generated using good quality dNTPs, enzymes and primers. Although using gel purification of PCR products to achieve specific PCR products is a common practice, PCR condition optimization is a better solution to avoid generation of multiple PCR products and further gel purification. Purified PCR products from the agarose gels may get contaminated with sequencing inhibitors. Instead, magnetic beads, such as ADS™ PCR Cleaning Magnetic Beads, are often suggested for cost-effective clean-up of the PCR products.
Another common used method for cleaning up of the PCR products, especially for Sanger sequencing and NGS, is an enzymatic method to degrade left-over PCR primers and dephosphorylate dNTPs. ADS Exo-Alp PCR Cleanup Mix, alternative to ExoSAP-IT PCR Product Cleanup Reagent, can be directly added to the PCR tubes to remove the interference of PCR primers and dNTPs in 30 minutes and the reactions can be directly used for downstream applications without further purification.
Amplification of plasmid template from single colonies
For mutation screening purposes, plasmid template can be amplified directly from single colonies without growing the culture for plasmid isolation. This is especially popular when only a small fraction of colonies contains the plasmid inserts with desired mutations. The plasmid amplification can be achieved through rolling circle amplification (RCA) using Bacillus subtilis phi29 DNA polymerase, degenerate primers, and little cell lysate from single colonies. The amplification products can be directly used for Sanger sequencing without further purification.
| Template Products |
| ADS PCR and Purification Kit |
ADS PCR and PCR Cleaning Beads |
ADS Exo-Alp PCR Cleanup Mix |
ADS phi29 DNA Polymerase and RCA DNA Amplification Kit |
| High quality for PCR success and cleaning |
For efficient contaminant removal |
For quick removal of PCR primers and dephosphorylation of dNTPs |
For fast sequencing of DNA from single colonies |