The first hands-on aspect in any RNA sequencing experiment is obtaining and properly storing the target sample for downstream analysis. Cell fixation followed by RNA isolation (including RNA stabilization) are arguably two of the most important steps in preparing for RNA sequencing, as they affect the quality of all downstream analyses and results.
Importance of Quality
In a publication which “sought to quantify the impact of variation in RNA quality on estimates of gene expression levels based on RNA-seq data” 1 the authors “observed widespread effects of RNA quality on measurements of gene expression levels, as well as a slight but significant loss of library complexity in more degraded samples.”1
Another publication calls out whole transcriptome amplification (WTA) steps, which it says “rely on additional PCR steps that are known to introduce amplification biases in the gene expression data.”2
So RNA quality is important, but how is it measured? And how can scientists effectively improve RNA quality for their experiments?
RNA Integrity Number (RIN)
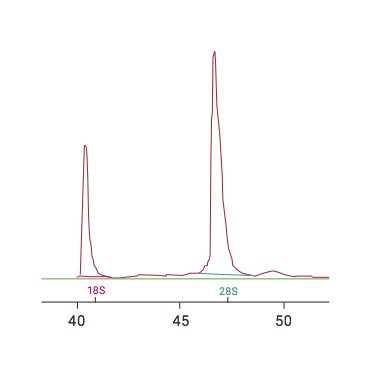
RNA quality was previously calculated using the ratio of 28S:18S ribosomal RNA with a spectrophotometer. This was shown to be fairly inconsistent.3
The RNA Integrity Number (RIN) is a now the widely accepted measure of RNA sample quality. It is an algorithm that uses electrophoretic RNA measurements recorded using microcapillary electrophoresis.3
“Although a higher RIN value is desirable, there is no definitive universally accepted RIN value for sample inclusion…Proposed thresholds for sample inclusion have varied between RIN values as high as 8 and as low as 3.95.”1
Using CellCover for Fixation of Cells as a RNA Stabilizer and RNA Shield
Fixation of cells is achieved with CellCover by liquid freezing – the metabolic state of cells is frozen without applying low temperatures. Synthesis and turnover of bio-molecules are both stalled. This includes instant disruption of cellular degradation pathways, making cell fixation by CellCover very fast. Chemical degradation is inhibited with CellCover during cell fixation as well, making it an excellent RNA stabilizer.
CellCover acts as an RNA shield, and RNA degradation is prevented by using CellCover as a RNA stabilizer. Even the integrity of high molecular weight RNA is protected (e.g. precurser rRNA, which are normally degraded after applying commonly used standards procedures). There is no need to rush your experiments, as you can safely store your sample in CellCover at 4°C and isolate RNA later for RNA sequencing.
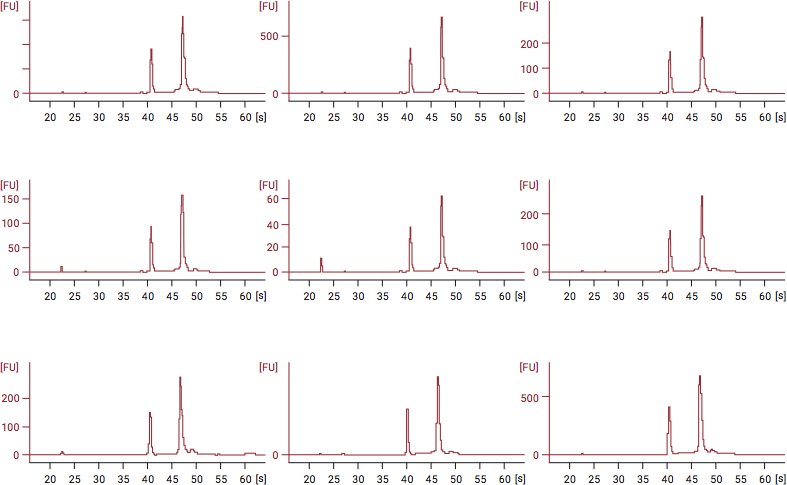
Below is an example of RIN analysis data after fixation of cells with CellCover for RNA stabilization. In the figures below, RNA was isolated and stored for up to 12 days at 4°C. The graphs depict an effective RNA shield, showing a consistent, stable RIN value of 10 from Day 1 through Day 12.