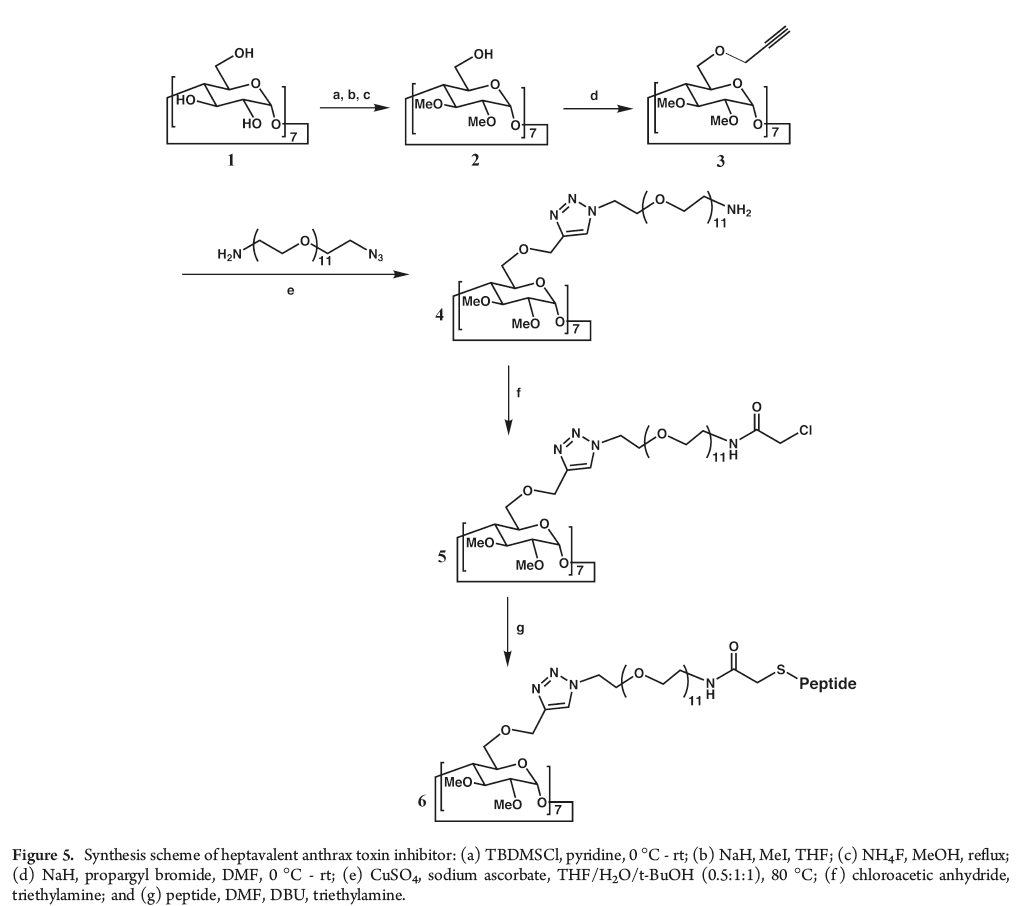
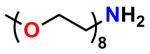Indeed, in testing the heptavalent anthrax toxin inhibitor, six out of seven rats treated with the inhibitor and exposed to anthrax did not develop anthrax. Conversely, rats exposed to anthrax and either not treated with inhibitor or treated with a sham inhibitor developed anthrax symptoms and died.
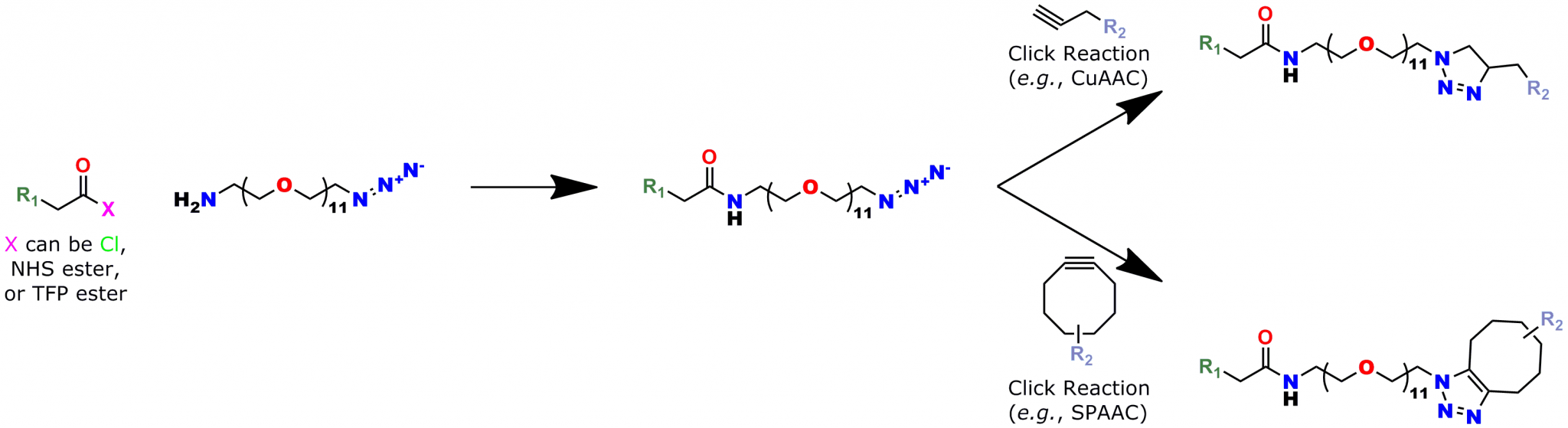
CuAAC is a simple reaction that gives high yields with minimal byproducts. This research shows the power of using click chemistry with a dPEG® reagent to exert spatial control over macromolecular product design. The precise spatial control needed to position the inhibiting peptide from the β-cyclodextrin core would not have been possible with a dispersed PEG. Only with a single molecular weight PEG (that is, a dPEG®) could that kind of control been obtained. Having a single molecular weight PEG product allows greater control over product design and purity. Also, compared to traditional, dispersed PEGs, a dPEG® compound simplifies product analysis. This research (among others) proves that point.
3. Atomic Force Microscopy
Atomic Force Microscopy (AFM) is a type of scanning probe microscopy with a resolution to fractions of a nanometer. One use of AFM measures the force between a probe and a sample. This use of AFM is known as force spectroscopy.
In single-molecule AFM force spectroscopy (SMFS), compounds of interest are attached by linkers to the cantilevers used to measure the force. Each cantilever attaches a single molecule (13), hence the name.
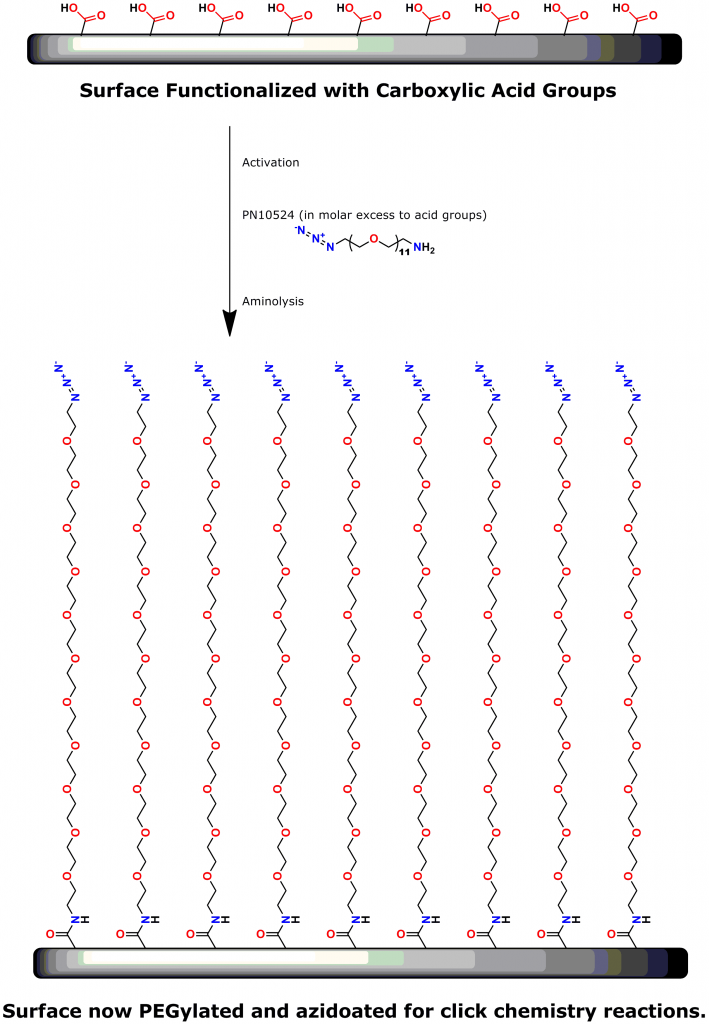
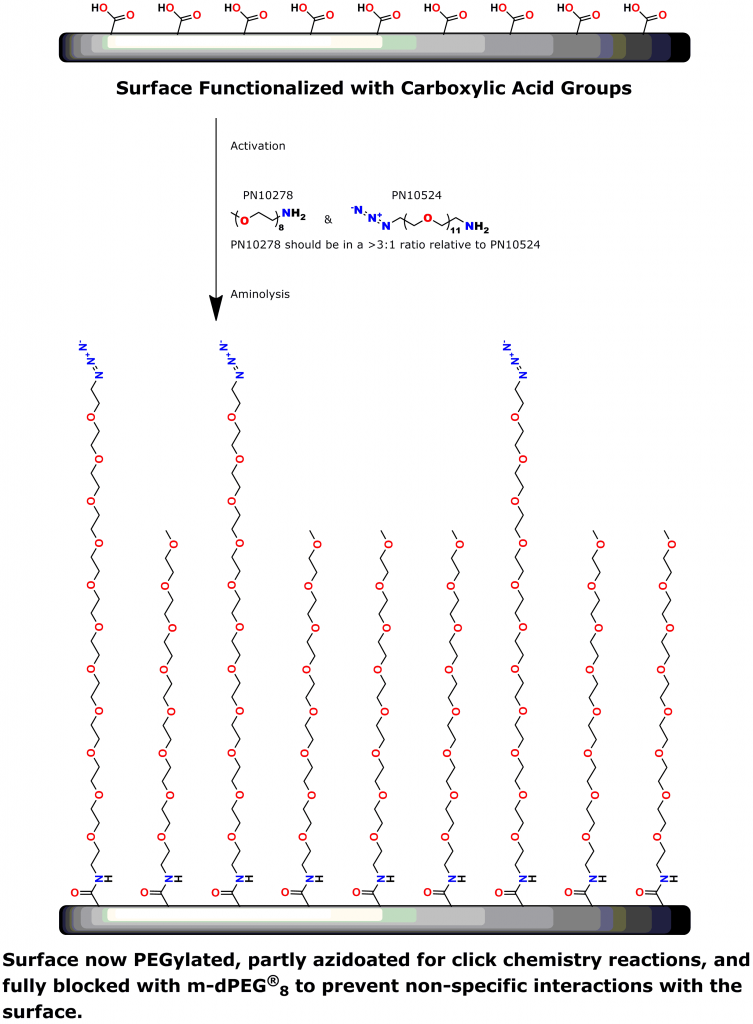
PEG is the most commonly used linker for attaching biomolecules to cantilevers (14-16). However, PEG polymers are dispersed and have varied chain lengths (15, 16). Thus, traditional PEG linkers are less than ideal for SMFS.
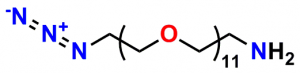
A 2012 Master’s Thesis by Jamie Maciaszek in the lab of Yuri L. Lyubchenko reported that linkers containing a single molecular weight and chain length were superior to traditional, dispersed PEG linkers (17). Moreover, using PN10524, azido-dPEG®11-amine, SMFS research in the lab of George Lykotrafitis detected and quantitatively mapped individual calcium-activated small conductance (SK) potassium channels in living neurons. The researchers joined the amine end of the crosslinker to APTES-activated silicon nitride cantilevers. Click chemistry crosslinking then linked the molecules of interest with the resulting azide-coated surface. The dPEG® linker was necessary for the research because it had no chain length heterogeneity (18).
Conclusion
Click chemistry crosslinking using Quanta BioDesign’s dPEG® products improves applications such as surface modification, macromolecular construction, and AFM (including SMFS). In this post, I showed how product number 10524, azido-dPEG®11-amine, is useful for all of these purposes. We make and sell this type of crosslinker in five sizes ranging from dPEG®3 (15.4 Å) to dPEG®35 (129.0 Å). Yo.
Buy the Products Mentioned
Please click the links below to go to the product pages of the products discussed in this post.
PN10278, m-dPEG®8-amine
PN10524, azido-dPEG®11-amine
References
1. Hartmuth C. Kolb, M. G. Finn, and K. Barry Sharpless. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chem. Int. Ed. (2001), 40, 2004-2021.
2. Hartmuth C. Kolb and K. Barry Sharpless. The Growing Impact of Click Chemistry on Drug Discovery. Drug Discovery Today (December 2003), 8(24), 1128-1137.
3. John E. Moses and Adam D. Moorhouse. The growing applications of click chemistry. Chem. Soc. Rev. (2007), 36, 1249-1262.
4. Christopher D. Hein, Xin-Ming Liu, and Dong Wang. Click Chemistry, a Powerful Tool for Pharmaceutical Sciences. Pharm Res (October 2008), 25(10), 2216-2230.
5. Charles E. Hoyle and Christopher N. Bowman. Thiol-Ene Click Chemistry. Angew. Chem. Int. Ed. (2010), 49(9), 1540-1573.
6. Charles E. Hoyle, Andrew B. Lowe, and Christopher N. Bowman. Thiol-click chemistry: a multifaceted toolbox for small molecule and polymer synthesis. Chem. Soc. Rev. (2010), 39, 1355-1387.
7. Chao-Jun Li and Barry M. Trost. Green chemistry for chemical synthesis. Proc. Nat. Acad. Sci. (September 9, 2008), 105(36), 13197-13202.
8. Greg T. Hermanson. “Chemoselective Ligation; Bioorthogonal Reagents,” in Bioconjugate Techniques, 3rd edition. New York: Academic Press, 2013, page 771. We at Quanta BioDesign recommend Greg’s book to all of our customers. You can buy it from us. To get started, please click this link and then click “Add to Cart” to order the book.
9. Gangadhar Jogikalmath. Method for blocking non-specific protein binding on a functionalized surface. US 20080213910 A1, September 4, 2008.
9a. Nick R. Glass, Ricky Tjeung, Peggy Chan, Leslie Y. Yeo, and James R. Friend. Organosilane deposition for microfluidic applications. Biomicrofluidics (2011), 5(3), 036501–036501-7. DOI: 10.1063/1.3625605. PMCID: PMC3364836.
10. Jacob Piehler, Andreas Brecht, Ramūnas Valiokas, Bo Liedberg , and Günter Gauglitz. A high-density poly(ethylene glycol) polymer brush for immobilization on glass-type surfaces. Biosensors & Bioelectronics (2000), 15, 473–481.
11. Hongwei Chen, Julie Yeh, Liya Wang, Xinying Wu, Zehong Cao, Y. Andrew Wang, Minming Zhang, Lily Yang, and Hui Mao. Reducing Non-Specific Binding and Uptake of Nanoparticles and Improving Cell Targeting with an Antifouling PEO-b-PγMPS Copolymer Coating.. Biomaterials (July 2010), 31(20): 5397–5407. DOI: 10.1016/j.biomaterials.2010.03.036
12. Amit Joshi, Sandesh Kate, Vincent Poon, Dhananjoy Mondal, Mohan B. Boggara, Arundhati Saraph, Jacob T. Martin, Ryan McAlpine, Ryan Day, Angel E. Garcia, Jeremy Mogridge, and Ravi S. Kane. Structure-Based Design of a Heptavalent Anthrax Toxin Inhibitor. Biomacromolecules (2011), 12(3), 791–796. DOI: 10.1021/bm101396u.
13. Keir C. Neuman and Attila Nagy. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nature Methods (June 2008), 5(6), 491-505. DOI: 10.1038/nmeth.1218
14. Bo-Hyun Kim, Nicholas Y Palermo, Sándor Lovas, Tatiana Zaikova, John Keana, and Yuri Lyubchenko. Single molecule atomic force microscopy force spectroscopy study of Aß-40 interactions. Biochemistry (2011), 50(23), 5154–5162. DOI: 10.1021/bi200147a.
15. Timothy V. Ratto, Kevin C. Langry, Robert E. Rudd, Rodney L. Balhorn, Michael J. Allen, Michael W. McElfresh. Force Spectroscopy of the Double-Tethered Concanavalin-A Mannose Bond. Biophysical Journal (2004), 86(4), 2430-2437.
16. Zenghan Tong, Andrey Mikheikin, Alexey Krasnoslobodtsev, Zhengjian Lv, Yuri L. Lyubchenko. Novel polymer linkers for single molecule AFM force spectroscopy. Methods (April 2013), 60(2), 161-168.
17. Jamie L. Maciaszek. Detection of SK2 Channels on Hippocampal Neurons. Master’s Thesis. University of Connecticut Graduate School, 2012.
18. Jamie L. Maciaszek, Heun Soh, Randall S. Walikonis, Anastasios V. Tzingounis, and George Lykotrafitis. Topography of Native SK Channels Revealed by Force Nanoscopy in Living Neurons. The Journal of Neuroscience (2012), 32(33), 11435-11440. DOI:10.1523/JNEUROSCI.1785-12.2012.