The inhibition of viral proteases necessary for proteolytic processing of polyproteins has been a successful strategy in the treatment of human immunodeficiency virus (HIV) and hepatitis C respectively, proving the potential of protease inhibitors for the treatment of viral infections. Similarly, the main protease of SARS-CoV-2 is thought to be essential for viral replication and, therefore, is regarded as promising target for antiviral therapy of Covid-19.
There is an extremely urgent need for the development of specific antiviral therapeutics and vaccines against SARS-CoV-2. The coronavirus main protease, which plays a pivotal role in viral gene expression and replication through the proteolytic processing of replicase polyproteins, is an attractive target for anti-CoV drug design.
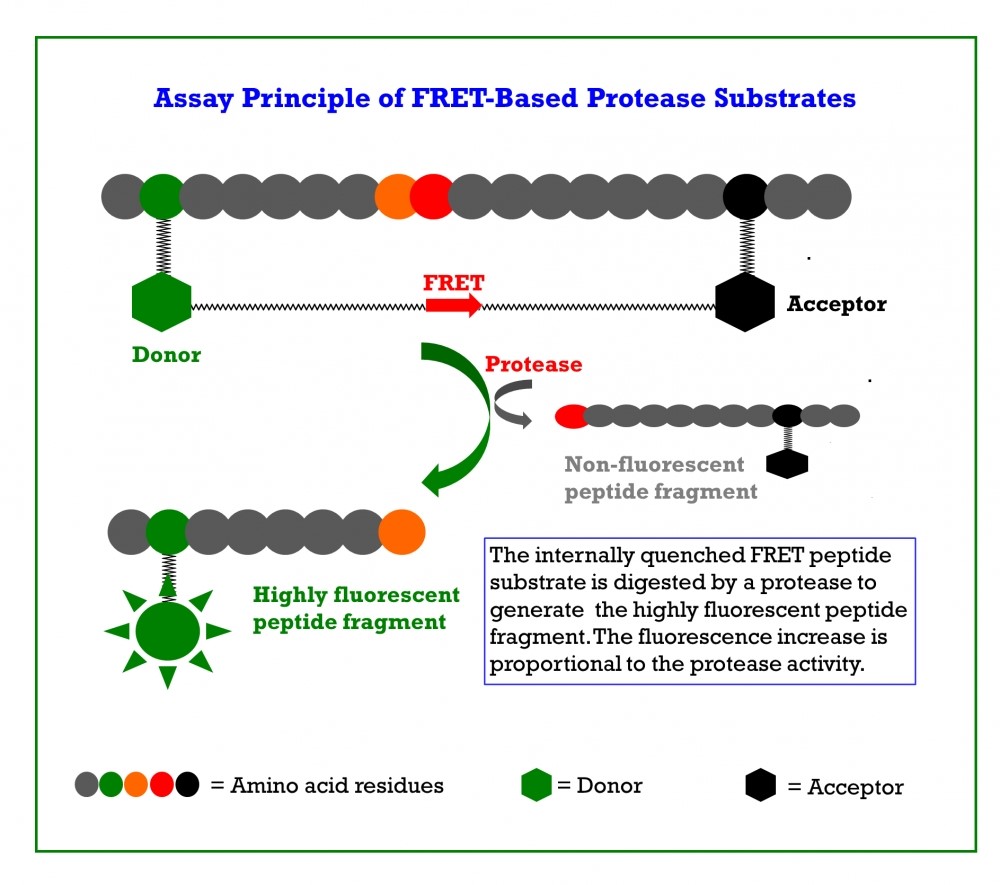
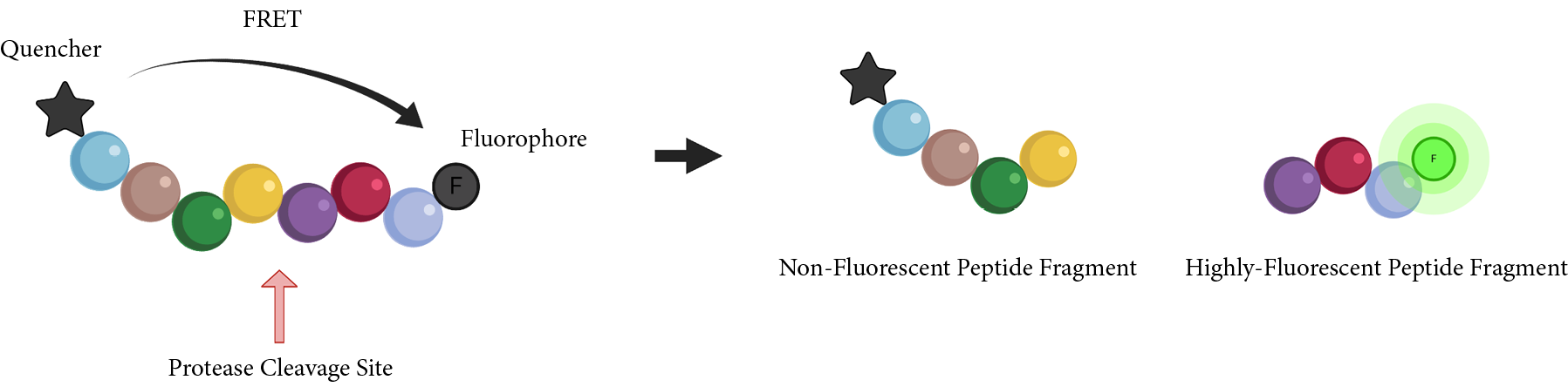
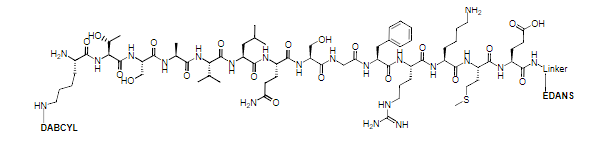
Multiple peptide substrates have been developed for monitoring the intensity of coronavirus proteases via FRET.
The Covidyte™ ED450, Covidyte™ EN450, Covidyte™ TF670, and Covidyte™ IF670 peptide substrates provide strong, extended fluorescent signal for searching inhibitors of coronavirus proteases with high sensitivity.
The papain-like proteases (PLpro) of coronaviruses carry out proteolytic maturation of non-structural proteins that play a role in replication of the virus and performs deubiquitination of host cell factors to scuttle antiviral responses. The newly-developed Covipyte™ EN450 is a peptide substrate containing the 9 amino acid sequence (RELNGGAPI) that can be cleaved by coronavirus PLpro, used for screening and studying the kinetics of PLpro inhibitors.
Additionally, Z-KAGG-AMC, Z-LRGG-AMC, and Z-KKAG-AMC are cleaved by papain-like proteases to give the highly fluorescent AMC product, to measure protease activity, and might be used for screening and studying kinetics of PLpro inhibitors.