Description
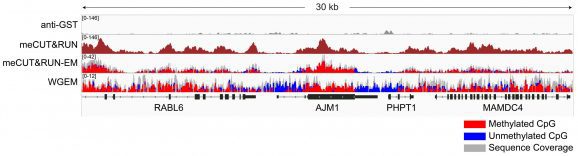
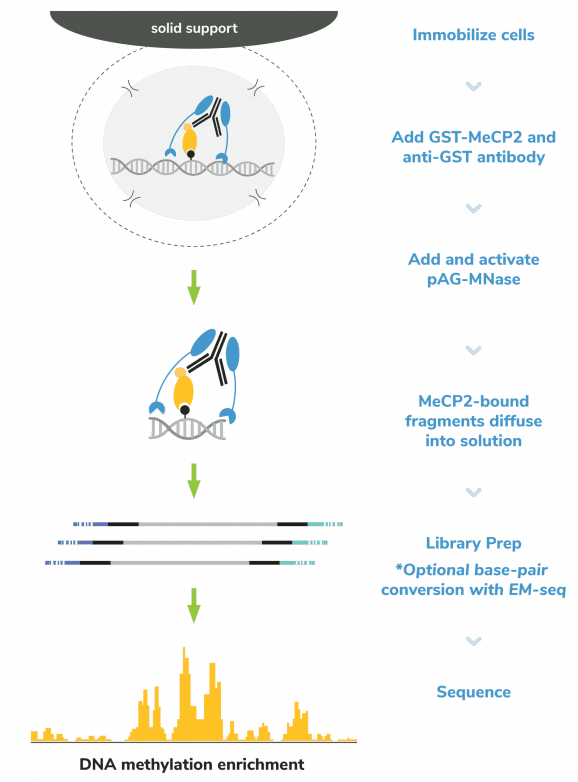
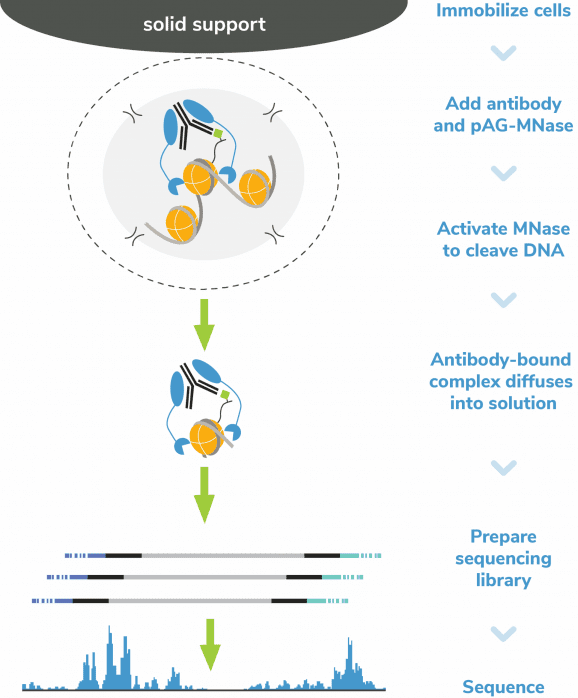
The CUTANA™ meCUT&RUN Kit for DNA Methylation Sequencing enables streamlined, high-resolution mapping of DNA methylation at >20-fold reduced sequencing depths compared to whole genome strategies like bisulfite sequencing. In meCUT&RUN, a GST-tagged MeCP2 methyl binding domain binds methylated DNA, directing the selective cleavage and release of DNA methylation-enriched chromatin fragments into solution by an immunotethered nuclease (pAG-MNase). Enriched DNA is separated from bead-bound cells, purified, and prepared for sequencing by one of two methods:
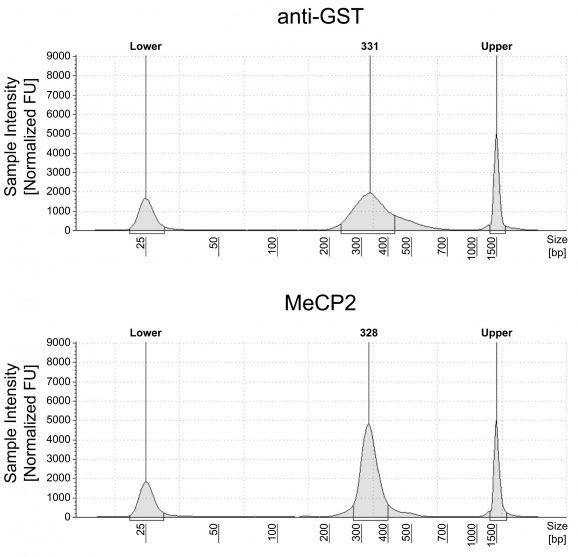
Option 1 uses a traditional library prep method, such as the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher 14-1001/14-1002), to provide ~150 bp resolution profiles of DNA methylation enrichment. Only 15-20 million total sequencing reads are required for this strategy, and data are analyzed using standard CUT&RUN bioinformatic pipelines.
Option 2 requires Enzymatic Methyl-seq (NEB® EM-seq™) or alternatively bisulfite sequencing to provide base-pair resolution of 5-methylcytosine (5mC). This workflow requires 30-50 million total sequencing reads and is analyzed using standard DNA methylation bioinformatic tools; see User Manual for detailed information.
The meCUT&RUN Kit is designed for 8-strip tubes and multi-channel pipetting, enabling a seamless workflow that maximizes throughput and reproducibility. The kit is compatible with a variety of inputs including cells or nuclei derived from native, cryopreserved, or cross-linked samples. While it is recommended to start with 500,000 cells, comparable data can be generated using as few as 10,000 cells. The low sequencing depth requirements, as well as compatibility with diverse sample inputs and low cell numbers, make this kit an ideal solution for DNA methylation sequencing.