Description
The CUTANA™ Multiomic CUT&RUN Controls Set is a specialized set of controls designed to be paired with the CUTANA™ ChIC/CUT&RUN Kit (EpiCypher 14-1048) to enable direct, simultaneous analysis of DNA methylation and chromatin proteins in a single workflow.
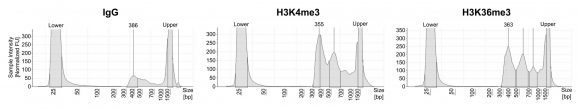
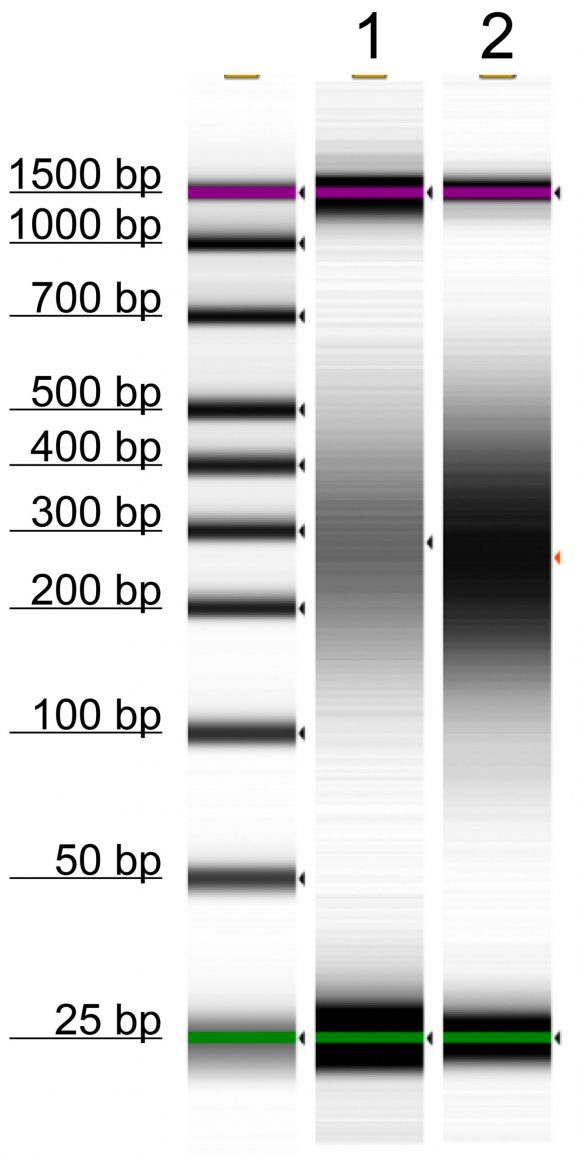
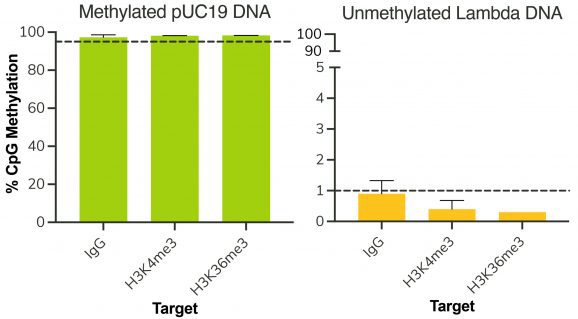
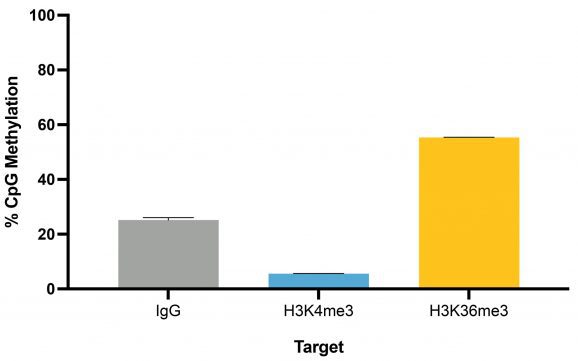
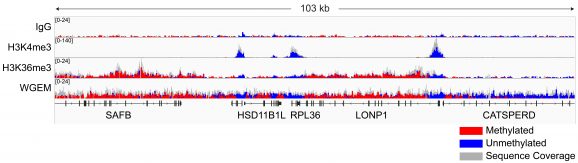
Following isolation of chromatin protein-bound DNA in CUT&RUN, DNA fragments are prepared for sequencing with a cytosine conversion strategy such as Enzymatic Methyl-seq (NEB® EM-seq™, preferred) or bisulfite sequencing to provide base-pair resolution of 5-methylcytosine (5mC). This set includes the essential controls for CUT&RUN followed by EM-seq (CUT&RUN-EM). H3K36me3 antibody is a positive control that enriches DNA associated with active gene bodies, which contain high levels of DNA methylation. H3K36me3 antibody complements the antibodies included in the CUT&RUN kit (H3K4me3, which enriches unmethylated promoters, and IgG, which serves as a negative control) to validate the technical success of the experimental workflow. Pre-fragmented methylated pUC19 and unmethylated Lambda DNAs serve as controls in EM-seq to assess conversion efficiency of unmethylated cytosines.
This controls set is a tailored solution to unlock the capacity of CUT&RUN to generate multiomic insights and investigate crosstalk between DNA methylation and chromatin proteins.