Epigenetics is one of the fastest-growing areas of academic and pharmaceutical research, with many projects focused on understanding the functions of histone post-translational modifications (PTMs) on chromatin. Indeed, manipulating or altering histone PTM interacting proteins, such as “readers” and/or chromatin modifying enzymes, is an attractive therapeutic strategy.
However, the precise PTM targets of many of these proteins is unknown. Notably, there are few reliable methods available to screen proteins against PTMs in a physiological, nucleosome context. These approaches also lack the broad substrate diversity and assay sensitivity needed to detect binding across a wide array of related / common PTMs.
dCypher™ Nucleosome Panels: Unparalleled Diversity for Rapid Histone PTM Profiling
dCypher™ : A Novel Platform to Interrogate Binding to PTMs
EpiCypher developed dCypher, a platform of both services and products with these exact needs in mind. Our dCypher services combine EpiCypher’s library of modified recombinant designer nucleosomes (dNucs) with AlphaScreen™ (Perkin Elmer™), a no-wash, bead-based proximity assay.

We discussed EpiCypher’s dCypher platform in a previous post, highlighting its functionality in delineating the specificity of reader proteins on histone peptide vs. dNucs. As shown in Figure 1, interaction of chromatin readers with dNucs brings the AlphaScreen Donor and Acceptor beads into close proximity, enabling light-induced excitation of the Acceptor bead, which is released in the form of fluorescence. Although a chromatin reader domain is shown here, this platform could be used to screen any number of chromatin-interacting proteins, including writers, erasers, and potential drugs or inhibitors.
New dCypher™ Nucleosome Panels Enable Rapid Screening of PTMs
EpiCypher has a diverse collection of modified dNucs, encompassing all of the most commonly studied PTMs in epigenetics research. We have recently released these dNucs in a series of panels in 96-well plate format, allowing their implementation for user-friendly, high-throughput discovery assays. Together our dCypher Nucleosome Panels and assay services provide scientists with ready access to nucleosomal PTM diversity to interrogate epigenetic regulation and associated diseases.
These panels come in four unique flavors, each offering a comprehensive set of related PTMs and histone variants (Figures 2 and 3). Three of our sets enable focused research on specific PTM families, notably histone lysine methylation + oncogenic histone mutations (Figure 2A), histone lysine acylation (Figure 2B), and histone arginine methylation (Figure 2C). For high-impact screens, we also offer a dCypher Nucleosome Full Panel, which contains 77 unique nucleosomes, including 60 modified dNucs, 8 onco-mutations (oncoNucs), 4 histone variants (vNucs), and 2 methylated DNA dNucs (MeDNA dNucs; Figure 3). Our panels provide truly unprecedented diversity for profiling chromatin interactor proteins (e.g. readers, enzymes, antibodies, etc.) against highly defined physiological nucleosome substrates.


Nucleosomal Context Alters Chromatin Reader Binding Specificity
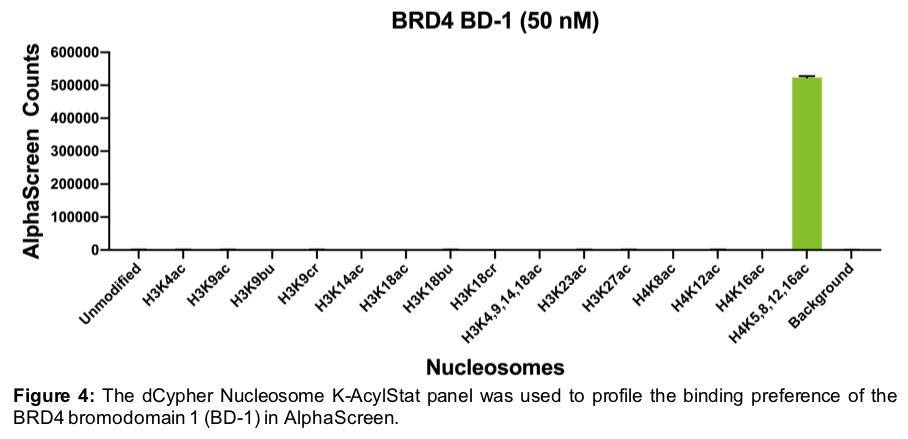
We have applied our dCypher Nucleosome Panels to a variety of chromatin interactors, including the BRD4 bromodomain 1 (BD-1; Figure 4). Interestingly, using our dCypher Nucleosome K-AcylStat panel we found that BRD4 (BD-1) shows high binding preference for the H4K5/8/12/16 poly-acetylated dNuc, but little interaction with single-acetylated or other acyl-modified dNucs in the panel. These results are in agreement with its reported preference on modified histone peptides1.
dCypher has also revealed novel, restricted specificity for chromatin readers on dNucs, versus binding on modified histones. As detailed in our previous blog post, using dCypher we found that the HP1β chromodomain only binds to H3K9me3 dNuc substrates, but binds to all three H3K9me1, me2, and me3 histone peptides. This is a recurring trend across our dCypher testing, and illustrates the power of the dCypher platform in defining chromatin reader binding interactions against a wide array of single and combinatorially-modified dNuc and histone peptide substrates.
As mentioned earlier, our dCypher Nucleosome panels can be utilized as substrates or controls for a variety of assay types, including AlphaScreen, immunoprecipitation, immunoblotting, ELISA, or custom user applications (the possibilities are endless!). Each dCypher plate comes with enough dNuc substrate to perform >35 AlphaScreen assays, >30 IP-qPCR experiments, and 3-5 immunoblotting tests.
If you are in need of a customized plate, or just want to learn more about the dCypher platform, drop us a line at info@stratech.co.uk.
REFERENCES
1. Filippakopoulos P, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell, 2012. 149(1): p. 214-31. (PubMed PMID: 22464331) (PMC3326523)