In a survey conducted by Nature, 90% of respondents agreed that there was a reproducibility crisis in biological experiments and many blamed it on poor-quality antibodies. Irreproducible results not only frustrate researchers, but also waste precious time, samples, and funding.
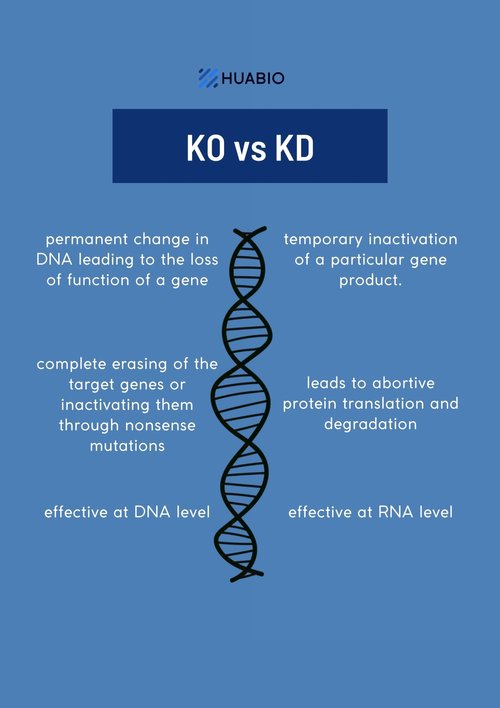
To validate an antibody, it must be shown to be specific, selective, and reproducible in the context for which it is to be used. One tried and true validation method is using the proper controls to ensure the absence of non-specific binding. For example, negative controls are particular samples included in the experiment that is treated the same as all the other samples but are not expected to change in any way due to the experimental conditions. The best negative control is a cell line or tissue that is known not to express the protein of interest. Testing antibody performance against genetically modified samples is one way to verify that an antibody recognizes a specific target. This can be done through various methods, two of which are knockdown and knockout samples.
What are Knockout samples?
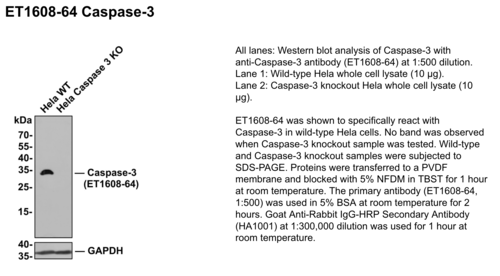
CRISPR/Cas9 gene-editing technology enables complete removal or “knock out” of both alleles of the gene encoding the target protein. Antibody specificity is confirmed by demonstrating that a protein band is only present in the wildtype and not the KO cell lysate in WB analysis.