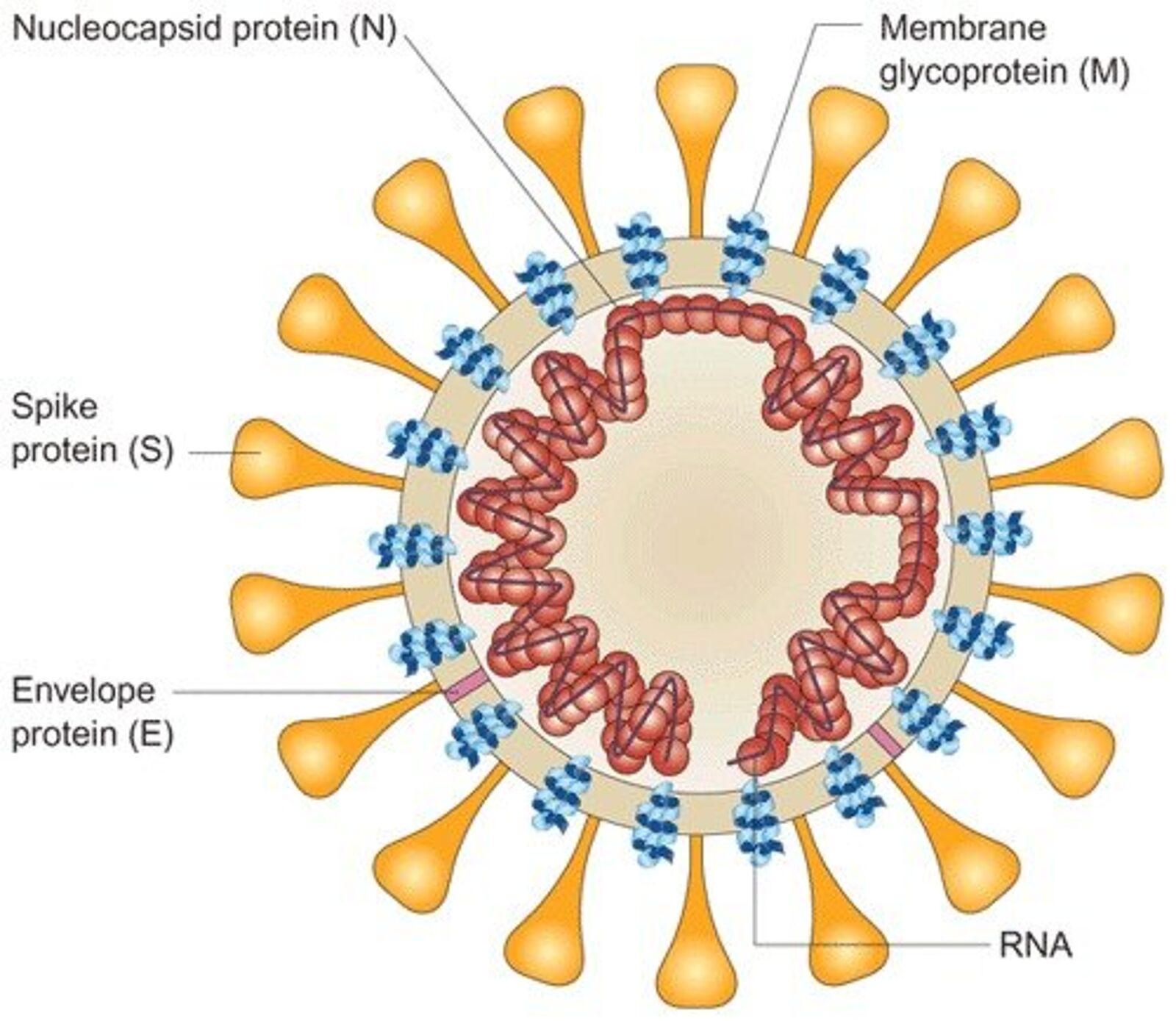
A SARS-CoV-2 virion is 50–200 nanometres in diameter. Like other coronaviruses, SARS-CoV-2 consists of four structural proteins, known as the S (spike), E (envelope), M (membrane), and N (nucleocapsid) proteins. The N protein holds the RNA genome, and the S, E, and M proteins together create the viral envelope. The spike protein is the protein responsible for allowing the virus to attach to and fuse with the membrane of a host cell: the S1 subunit catalyzes attachment, the S2 subunit fusion.
LUBIO COVID-19
Research Solutions for Coronavirus – Lubio Science
Available now: SARS-CoV-2 full-length trimeric spike protein
Full-length trimeric spike protein from SARS-CoV-2 coronavirus, the causative agent of COVID-19. Available as highly purified his-tagged trimeric protein in solution.
| SARS-CoV-2 trimeric spike protein (wt) | SARS-CoV-2 trimeric spike protein (D614G) | |
| Conformation | Wild type trimeric SARS-CoV-2 spike protein in prefusion conformation | D614G variant trimeric SARS-CoV-2 spike protein in prefusion conformation |
| Modifications | C-terminal Transmembrane region replaced with a trimerization domain and a polyhistidine tag. Two stabilizing proline mutations and scrambled S1/S2 furin cleavage site | C-terminal Transmembrane region replaced with a trimerization domain and a polyhistidine tag. Two stabilizing proline mutations. Scrambled S1/S2 furin cleavage site. D614G mutation |
| Strain | SARS-CoV-2 Betacoronavirus | SARS-CoV-2 Betacoronavirus |
| Isolate (Seq ID) | Wuhan-Hu-1 (GenBank: MN908947) | Wuhan-Hu-1 (GenBank: MN908947) D614G variant |
| Expression System | CHOExpressTM cells | CHOExpressTM cells |
| Purity | > 90 % as determined by SDS-PAGE | > 90 % as determined by SDS-PAGE |
| Buffer | 0.01M PBS, pH 7.4 , no preservatives | 0.01M PBS, pH 7.4 , no preservatives |
| Datasheet |
| Cat-No. | Item | Size | Price |
| LU2010-50UG | Trimeric spike protein from SARS-CoV-2 | 50 ug | £545 |
| LU2011-50UG | Trimeric spike protein from SARS-CoV-2, D614G variant | 50 ug | £545 |
VARIANTS OF SARS-COV-2 FULL-LENGTH TRIMERIC SPIKE RECOMBINANT PROTEIN
Full-length trimeric spike variants from SARS-CoV-2, including the full transmembrane domain as well as full glycosylation markers.



| SARS-CoV-2 full-length Trimeric Spike Recombinant Protein (UK Variant) | SARS-CoV-2 full-length Trimeric Spike Recombinant Protein (South African Variant) | SARS-CoV-2 full-length Trimeric Spike Recombinant Protein (Brazil Variant) | |
| Description | Spike protein of the mutant strain B.1.1.7, also commonly known as the “UK Variant”. It is a full-length protein, which is active in its native trimeric form, that is stabilized in LMNG detergent. | Spike protein of the mutant strain B.1.351, also commonly known as the “South Africa Variant”. It is a full-length protein, which is active in its native trimeric form, that is stabilized in LMNG detergent. | Spike protein of the mutant strain P.1, also commonly known as the “Brazil Variant”. It is a full-length protein, which is active in its native trimeric form, that is stabilized in LMNG detergent. |
| WHO reference | SARS-CoV-2 VUI 202012/01 | 501Y.V2/501.V2 | B.1.128 |
| Mutations | del 69-70, del 144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | del 144, K417N, E484K, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, H655Y, T1027I, V1176F |
| Further modifications | -Furin cleavage site “RRAR” mutated to “GSAG”; KV986PP | Further modifications | -Furin cleavage site “RRAR” mutated to “GSAG”; KV986PP |
| -C-terminal Rho1D4 tag fused with spacer “GSSG” to protein sequence | -Furin cleavage site “RRAR” mutated to “GSAG”; KV986PP | -C-terminal Rho1D4 tag fused with spacer “GSSG” to protein sequence | -Furin cleavage site “RRAR” mutated to “GSAG”; KV986PP |
| -C-terminal Rho1D4 tag fused with spacer “GSSG” to protein sequence | -Furin cleavage site “RRAR” mutated to “GSAG”; KV986PP | -C-terminal Rho1D4 tag fused with spacer “GSSG” to protein sequence | -Furin cleavage site “RRAR” mutated to “GSAG”; KV986PP |
| -C-terminal Rho1D4 tag fused with spacer “GSSG” to protein sequence | -C-terminal Rho1D4 tag fused with spacer “GSSG” to protein sequence | -C-terminal Rho1D4 tag fused with spacer “GSSG” to protein sequence | -C-terminal Rho1D4 tag fused with spacer “GSSG” to protein sequence |
| Expression System | HEK293 | HEK293 | HEK293 |
| Tag | C-terminal Rho1D4 | C-terminal Rho1D4 | C-terminal Rho1D4 |
| Purity | >98% as determined by SDS-PAGE, see datasheet | >98% as determined by SDS-PAGE, see datasheet | >98% as determined by SDS-PAGE, see datasheet |
References
- Cryo-EM Structure of the 2019-nCoV Spike in the Prefusion Conformation (LINK TO PUBLICATION)
- Site-specific analysis of the SARS-CoV-2 glycan shield (LINK TO PUBLICATION)
- Structure-based design of prefusion-stabilized SARS-CoV-2 spikes (LINK TO PUBLICATION)
- Continuous flexibility analysis of SARS-CoV-2 Spike prefusion structures (LINK TO PUBLICATION)
- Broad sarbecovirus neutralizing antibodies define a key site of vulnerability on the SARS-CoV-2 spike protein (LINK TO PUBLICATION)
- Potent neutralizing antibodies directed to multiple epitopes on SARS-CoV-2 spike (LINK TO PUBLICATION)
- A pH-dependent switch mediates conformational masking of SARS-CoV-2 spike (LINK TO PUBLICATION)
