CUTANA™️Hia5 for Fiber-seq
Catalogue Number: 15-1032-EPC
| Manufacturer: | EpiCypher |
| Shelf Life: | 6 months |
| Molecular Weight: | 33.8 kDa |
| Type: | Transferases |
| Host Cell: | E. Coli |
| Shipping Condition: | Blue Ice |
| Storage Condition: | -80°C |
| Unit(s): | 8 Reactions, 24 Reactions |
| Host name: | |
| Clone: | |
| Isotype: | |
| Immunogen: | |
| Range: | |
| Sensitivity: | |
| Sample type: | |
| Sample size: |
Description
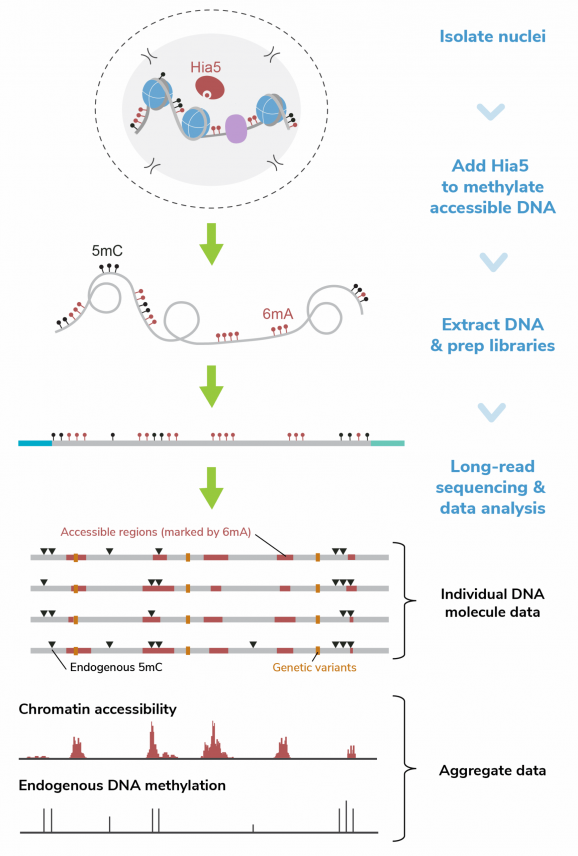
Description: CUTANA™ Hia5 is the key enzyme for Fiber-seq [1], a multiomic long-read sequencing (LRS) assay that simultaneously profiles chromatin accessibility, DNA methylation, and genetic variation at single-molecule resolution. By layering chromatin accessibility on top of standard LRS workflows, Fiber-seq uniquely links epigenetic information (DNA methylation) and complex genetic architecture (e.g., structural variants and haplotype phasing) to chromatin states within the same molecules of DNA.
Hia5 is a DNA adenine methyltransferase (MTase) that uses S-adenosylmethionine (SAM) to catalyze methylation at the N6 position of adenine, producing N6-methyladenine (6mA). Fiber-seq leverages the nonspecific activity of Hia5 to methylate adenines within accessible chromatin regions in situ. This records chromatin accessibility information onto native DNA while preserving endogenous methylation (5mC). Hia5-modified DNA is fully compatible with LRS on both Pacific Biosciences® (PacBio® HiFi Sequencing) and Oxford Nanopore Technologies® (ONT® Nanopore Sequencing) platforms.
Key Advantages
Multiomic insights in one assay – Fiber-seq simultaneously profiles chromatin accessibility, DNA methylation, and genetic variants, consolidating what typically requires three or more separate assays into one long-read sequencing assay.
Highest activity 6mA-MTase – Faster labeling via Hia5 enables higher resolution of accessible DNA, with demonstrated utility in protein footprinting applications to resolve transcription factor binding and nucleosome positioning [2-4].
Access complex genomic regions – Profile chromatin accessibility in repetitive and structurally complex regions such as transposable elements, centromeres, telomeres, segmental duplications, and chromosomal rearrangements.
Easy workflow integration – Fiber-seq can be performed in <2 hours directly upstream of routine long-read sequencing library preparation.
Resolve chromatin state heterogeneity – PCR-free approach leverages long-read sequencing to directly profile individual DNA molecules, uncovering chromatin structure heterogeneity in cell populations often missed by short-read techniques.
Background References:
[1] Stergachis et al. Science (2020). PMID: 32587015
[2] Tullius et al. Mol. Cell (2024). PMID: 39191261
[3] Dubocanin et al. Cell Genom. (2025). PMID: 40147439
[4] Vollger et al. bioRxiv (2024). DOI: 10.1101/2024.06.14.599122
[5] Jha et al. Genome Res. (2024). PMID: 38849157