A protease is an enzyme that conducts proteolysis, i.e., the protein catabolism by hydrolysis of the peptide bonds that link amino acids together in the polypeptide chain which form the protein. Proteases, also known as peptidases or proteolytic enzymes, are a large group of enzymes. They belong to the class of enzymes known as hydrolases, which catalyse the reaction of hydrolysis of various bonds with the participation of a water molecule.
Proteases
Major Protease Categories
Proteases are divided into four major groups according to the character of their catalytic active site and conditions of action. These groups include: serine proteinases, cysteine (thiol) proteinases, aspartic proteinases, and matrix metalloproteinases (MMPs). Attachment of a protease to a certain group depends on the structure of catalytic site and the amino acid (as one of the constituents) essential for its activity. Proteases are involved in digesting long protein chains into short fragments, splitting the peptide bonds that link amino acid residues. Some of them can detach the terminal amino acids from the protein chain (exopeptidases, such as aminopeptidases, carboxypeptidase A), while others attack internal peptide bonds of a protein (endopeptidases, such as trypsin, chymotrypsin, pepsin, papain and elastase).
Enzymatic Function and Mechanism
Proteases occur in all organisms. These enzymes are involved in a multitude of physiological reactions from simple digestion of food proteins to highly regulated cascades (e.g., the blood-clotting cascade, the complement system, apoptosis pathways, and the invertebrate prophenoloxidase-activating cascade). Proteases can either break specific peptide bonds, depending on the amino acid sequence of a protein, or break down a complete peptide to amino acids. The activity can be a destructive change, abolishing a protein’s function or digesting it to its principal components. It can be an activation of a function, or it can be a signal in a signaling pathway.
Metabolic Proteases
Proteases are used throughout an organism for various metabolic processes. Acid proteases secreted into the stomach (such as pepsin) and serine proteases present in duodenum (trypsin and chymotrypsin) enable us to digest the protein in food. Proteases present in blood serum (thrombin, plasmin, Hageman factor, etc.) play an important role in blood-clotting, as well as blood clot lysis, and the correct action of the immune system. Other proteases are present in leukocytes (elastase and cathepsin G) and play several different roles in metabolic control.
Proteases determine the lifetime of other proteins playing an important physiological role like hormones, antibodies, or other enzymes—this is one of the fastest “switching on” and “switching off” regulatory mechanisms in the physiology of an organism. By complex cooperative action the proteases may proceed as cascade reactions, which result in rapid and efficient amplification of an organism’s response to a physiological signal.
Biodiversity of Proteases
Bacteria also secrete proteases to hydrolyze (digest) the peptide bonds in proteins and therefore break the proteins down into their constituent monomers (amino acids). Bacterial and fungal proteases are particularly important to the global carbon and nitrogen cycles in the recycling of proteins, and such activity tends to be regulated by nutritional signals in these organisms. The net impact of nutritional regulation of protease activity among the thousands of species present in soil can be observed at the overall microbial community level as proteins are broken down in response to carbon, nitrogen, or sulfur limitation. A secreted bacterial protease may also act as an exotoxin, and be an example of a virulence factor in bacterial pathogenesis. Bacterial exotoxic proteases destroy extracellular structures. Protease assays are widely used for the investigation of protease inhibitors and the detection of protease activities. Monitoring various protease activities has become a routine task for many biological laboratories. Some proteases have been identified as good drug development targets.
Universal Protease Activity Assay
Amplite™ Universal Fluorimetric Protease Activity Assay Kit is an ideal choice to perform routine protease isolation assays or for identifying the presence of contaminating proteases in protein samples. The assay utilizes a fluorescent casein conjugate that is proven to be a generic substrate for a broad spectrum of proteases (e.g. trypsin, chymotrypsin, thermolysin, proteinase K, protease XIV, and elastase). The intact casein substrate is heavily labeled with a green fluorescent dye, resulting in significant fluorescence quenching. Protease-catalyzed hydrolysis relieves its quenching effect, yielding brightly fluorescent dye-labeled short peptides. The increase in fluorescence intensity is directly proportional to protease activity, and can be easily read with a fluorescence microplate reader at Ex/Em = 490/525 nm using the FITC filter set. The assay can be performed in a convenient 96-well or 384-well microtiter plate format and readily adapted to automation.
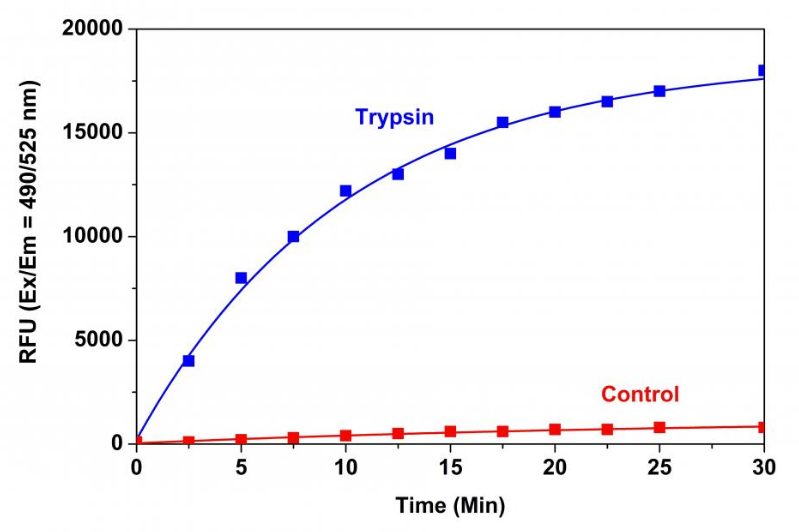
Figure 1. Trypsin protease activity was analyzed by Amplite™ Universal Fluorimetric Protease Activity Assay Kit. Protease substrate was incubated with 1 unit trypsin in the kit assay buffer. The control wells had protease substrate only (without trypsin). The fluorescence signal was measured starting from time 0 when trypsin was added. Samples were done in triplicates.
Table 1. Amplite™ Universal Fluorimetric Protease Activity Assay For Universal Protease Activity Assay
Matrix Metalloproteinases (MMPs)
Matrix metalloproteinases (MMPs) are a family of metalloproteinases that are calcium-dependent zinc-containing endopeptidases. Collectively, these enzymes are responsible for the degradation of most extracellular matrix proteins during growth and normal tissue turnover (e.g. breakdown of connective tissues, bone remodeling, the menstrual cycle and repair of tissue damage). MMPs are also key regulators of cell behavior and signaling pathways such as cell proliferation, migration, differentiation and apoptosis. They are known to be involved in the cleavage of cell surface receptors, the release of apoptotic ligands, and cytokine inactivation. While the exact contribution of MMPs to certain pathological processes is difficult to assess, MMPs appear to play a key role in the development of arthritis as well as in the invasion and metastasis of cancer.
Universal Matrix Metalloproteinase Activity Assays
Amplite™ Universal Fluorimetric MMP Activity Assay kits utilize a fluorescence resonance energy transfer (FRET) peptide as a generic MMP activity indicator. They can also be used to screen for MMP inhibitors when using purified MMP enzymes. In the intact FRET peptide, the fluorescence of one component is quenched by another. After cleavage into two separate fragments by MMPs, the fluorescence is recovered. With excellent fluorescence quantum yields and longer wavelengths, the FRET probe provided in each assay is much more sensitive than an EDANS/Dabcyl FRET substrate. The probe signal can be easily read by a fluorescence microplate reader at Ex/Em = 490/525 nm (Cat No. 13510) of at Ex/Em = 540/590 nm (Cat No. 13511).
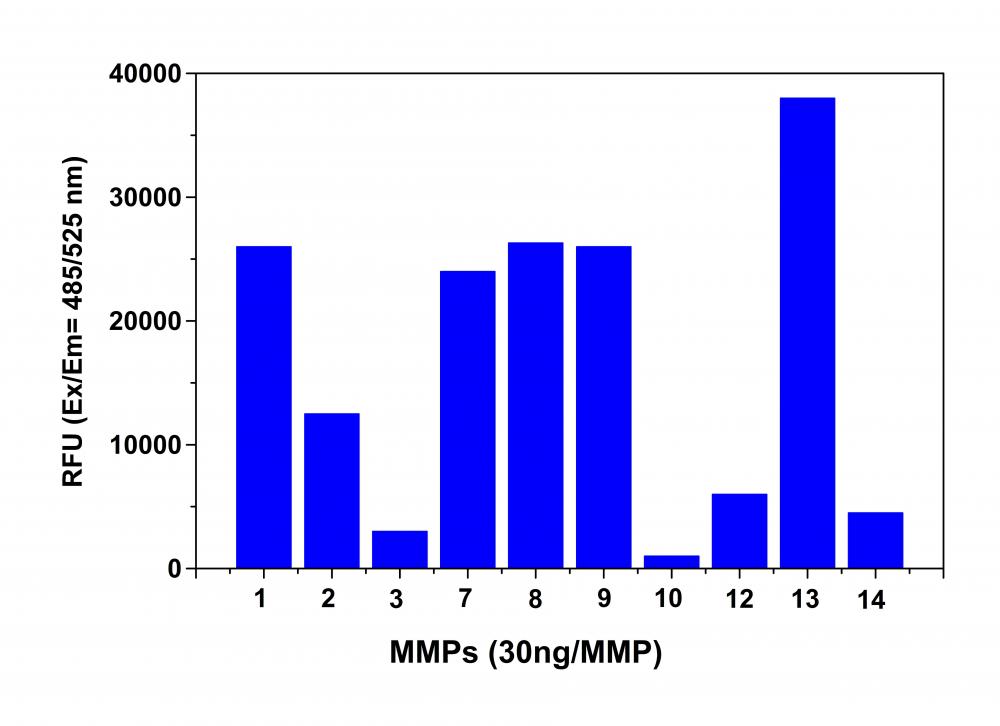
Figure 2. MMP activity monitored using Amplite™ Universal Fluorimetric MMP Activity Assay Kit. APMA-activated MMPs (30 ng each) were mixed with MMP Green™ substrate. One hour after starting the reaction the fluorescence signal was monitored using a NOVOStar microplate reader (BMG Labtech) at Ex/Em = 490/525 nm. The reading from all wells was subtracted by the reading from substrate control, which contains MMP Green™ substrate but no MMPs. The MMP Green™ substrate can detect the activity of sub-nanogram of all MMPs (n=3).
Table 2. Universal Matrix Metalloproteinase Activity Assays For Matrix Metalloproteinases (MMPs)
| Product Name | Mol. Wt. | Ex/Em (nm) | Donor Chromophore | Acceptor Chromophore |
| MMP Green™ Substrate | ∼2000 | 494/515 | Tide Fluor™ 2 | Tide Quencher™ 2 |
| MMP Red™ Substrate | ∼2000 | 543/570 | Tide Fluor™ 3 | Tide Quencher™ 3 |
Matrix Metalloproteinase-3
Amplite™ Fluorimetric MMP-3 Activity Assay
Amplite™ Fluorimetric MMP-3 Activity Assay Kit provides a quick and sensitive method to measure MMP-3 enzyme activity, as well as, screen for MMP-3 inhibitors. This assay utilizes a fluorescence resonance energy transfer (FRET) peptide sequence that is highly selective for MMP-3 hydrolysis over other MMP enzymes. In the intact FRET peptide, the fluorescence of Tide Fluor™ 2 (TF2) is quenched by Tide Quencher™ 2. After cleavage into two separate fragments by MMP-3 hydrolysis, the green fluorescence of TF2 is recovered. With excellent fluorescence quantum yield and longer wavelength, TF2 shows less interference from autofluorescence of test compounds and cellular components, and is much more sensitive than an EDANS/Dabcyl FRET substrate. The assay can be performed in a convenient 96-well or 384-well microtiter-plate format. The perfect excitation of TF2 at 488 nm makes the assay readily readable with almost all the common fluorescence instruments equipped with argon laser and FITC filter set.
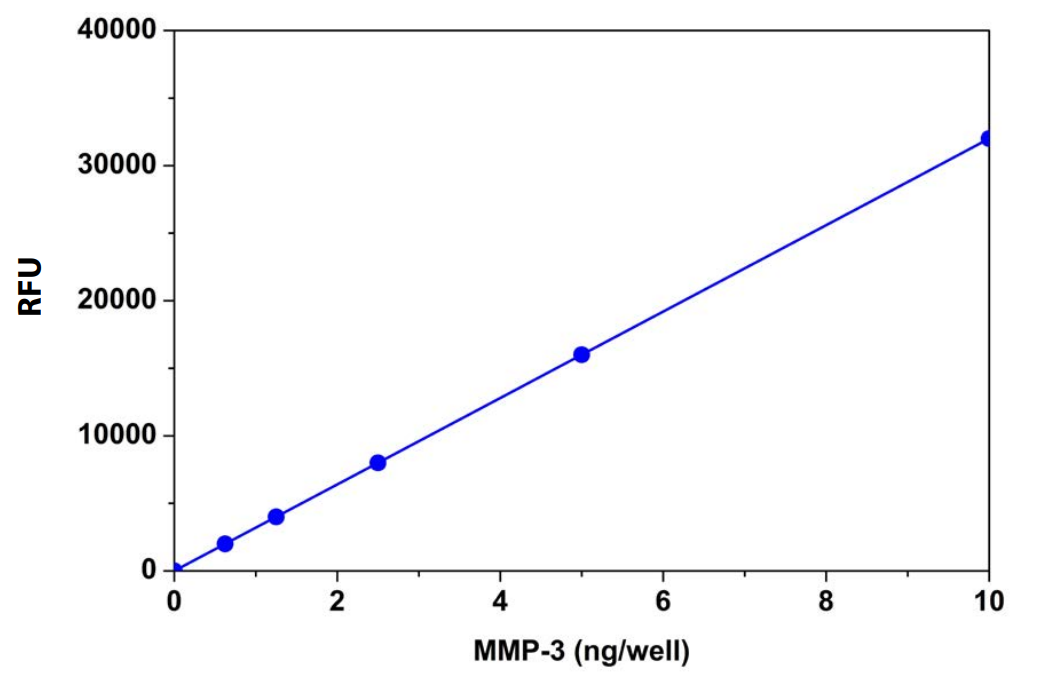
Table 3. Amplite™ Fluorimetric MMP-3 Activity Assay For Matrix Metalloproteinase-3
| Product Name | Mol. Wt. | Ex/Em (nm) | Donor Chromophore | Acceptor Chromophore |
| MMP-3 Green™ Substrate | ∼2000 | 494/515 | Tide Fluor™ 2 | Tide Quencher™ 2 |
Proteasome 20S
Proteasomes are protein complexes inside all eukaryotes, archaea, and in some bacteria. In eukaryotes, proteasomes are located in the nucleus and cytoplasm. The main function of the proteasome is to degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds. Enzymes that carry out such reactions are called proteases.
Function
Proteasomes are part of a major mechanism by which cells regulate the concentration of particular proteins and degrade misfolded proteins. The degradation process yields peptides of about seven to eight amino acids long, which can then be further degraded into shorter amino acid sequences and used in synthesizing new proteins. Proteins are tagged for degradation with a small protein called ubiquitin. The tagging reaction is catalyzed by enzymes called ubiquitin ligases. Once a protein is tagged with a single ubiquitin molecule, this is a signal to other ligases to attach additional ubiquitin molecules. The result is a polyubiquitin chain that is bound by the proteasome, allowing it to degrade the tagged protein.
Structure
In structure, the proteasome is a cylindrical complex containing a “core” of four stacked rings forming a central pore. Each ring is composed of seven individual proteins. The inner two rings are made of seven ? subunits that contain three to seven protease active sites. These sites are located on the interior surface of the rings, so that the target protein must enter the central pore before it is degraded. The outer two rings each contain seven ? subunits whose function is to maintain a “gate” through which proteins enter the barrel. These ? subunits are controlled by binding to “cap” structures or regulatory particles that recognize polyubiquitin tags attached to protein substrates and initiate the degradation process. The overall system of ubiquitination and proteasomal degradation is known as the ubiquitin-proteasome system.
Amplite™ Fluorimetric Proteasome 20S Activity Assay
The most common form of the proteasome in this pathway is the 26S proteasome, an ATP-dependent proteolytic complex, which contains one 20S (700-kDa) core particle structure and two 19S (700-kDa) regulatory caps. The 20S core contains three major proteolytic activities including chymotrypsin-like, trypsin-like and caspase-like activities. It is responsible for the breakdown of the key proteins involved with apoptosis, DNA repair, endocytosis, and cell cycle control.
Amplite™ Fluorimetric Proteasome 20S Activity Assay Kit is a homogeneous assay that measures the chymotrypsin-like protease activity associated with the proteasome complex in cultured cells. This kit uses LLVY-R110 as a fluorogenic indicator for proteasome activities. Cleavage of LLVY-R110 by proteasome generates strongly green fluorescent R110 which can be easily monitored by a fluorescence microplate reader at Ex/Em = 490/525 nm. The assay is robust and can be readily adapted for high-throughput assays to evaluate the proteasome activities or screen the inhibitors in cultured cells or in solution. The assay can be performed in a convenient 96-well and 384-well fluorescence microtiter-plate format
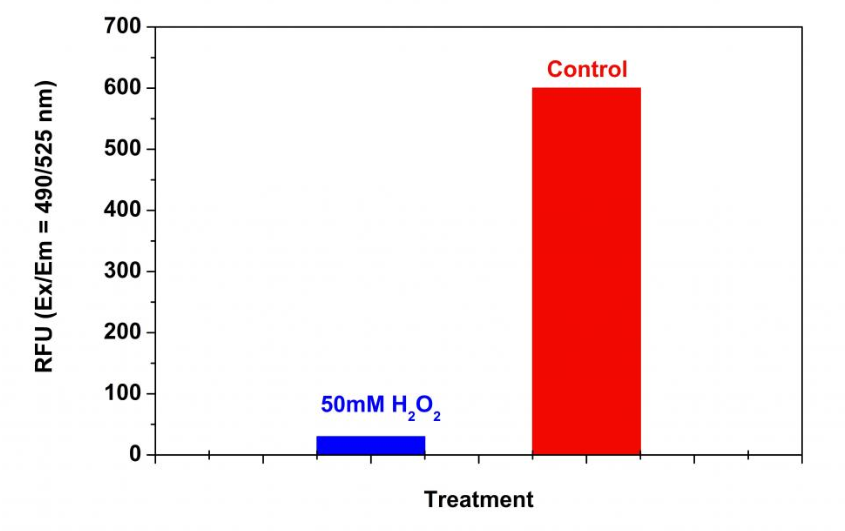
Figure 3. Detection of proteasome activity in Jurkat cells with Amplite™ Fluorimetric Proteasome 20S Activity Assay Kit. Jurkat cells were seeded on the same day at 500,000 cells/90 µL/well in a 96-well black wall/clear bottom Costar plate. The cells were treated with or without 50 mM H2O2 for 30 minutes. The proteasome assay loading solution (100 µL/well) was added and incubated in a 5% CO2, 37 °C incubator for 3 hours. The fluorescence intensity was measured at Ex/Em = 490/525 nm using a Gemini fluorescent microplate reader (Molecular Devices).
Table 4. Amplite™ Fluorimetric Proteasome 20S Activity Assay For Proteasome 20S
| Product Name | Mol. Wt. | Ex (nm) | Em (nm) | ε (cm-1M-1) | Function |
| (Ac-ANW)2R110 | 1159.16 | 499 | 521 | 80,000 | Proteasome 20S-β5i |
| (Ac-KQL)2R110 | 1153.33 | 499 | 521 | 80,000 | Proteasome 20S-β2i |
| (Ac-PAL)2R110 | 977.11 | 499 | 521 | 80,000 | Proteasome 20S-β1i |
| (Ac-WLA)2R110 | 1155.3 | 499 | 521 | 80,000 | Proteasome 20S-β5c |
| Suc-LLVY-AMC | 763.88 | 339 | 439 | N/A | Proteasome 20S-β5c |
| Suc-LLVY-Aminoluciferin | 868.03 | 360 | 496 | 18,000 | Proteasome 20S-β5c |
| (Suc-LLVY)2R110 | 1507.72 | 499 | 521 | 80,000 | Proteasome 20S-β5c |
| (Z-LLE)2R110 | 1309.46 | 499 | 521 | 80,000 | Proteasome 20S-β1c |
Renin
Renin is an enzyme that participates in the body’s renin-angiotensin system (RAS) that mediates extracellular volume and arterial vasoconstriction. It regulates blood pressure and electrolyte homoeostasis. Angiotensin II constricts blood vessels leading to increased blood pressure. It also increases the secretion of ADH and aldosterone, and stimulates the hypothalamus to activate the thirst reflex. An over-active renin-angiotension system leads to vasoconstriction and retention of sodium and water. Renin has been identified to be an attractive target for the treatment of hypertension.
Amplite™ Fluorimetric Renin Assay
Amplite™ Fluorimetric Renin Assay Kit provides a quick and convenient assay for high throughput screening of renin inhibitors and renin activity. The assay utilizes a fluorescence resonance energy transfer (FRET) peptide modified with the FRET pair Tide Fluor™ 3 (TF3) and Tide Quencher™ 3 (TQ3). In the intact FRET peptide, the fluorescence of TF3 is quenched by TQ3. Upon cleavage into two separate fragments by renin, the fluorescence of TF3 is recovered, and the fluorescence signal can be easily monitored by a fluorescence microplate reader at Ex/Em = 540/590 nm. This assay is approximately fifty fold more sensitive than an EDANS/Dabcyl-based assay, detecting as little as 1 ng renin in a 100 µL reaction volume.
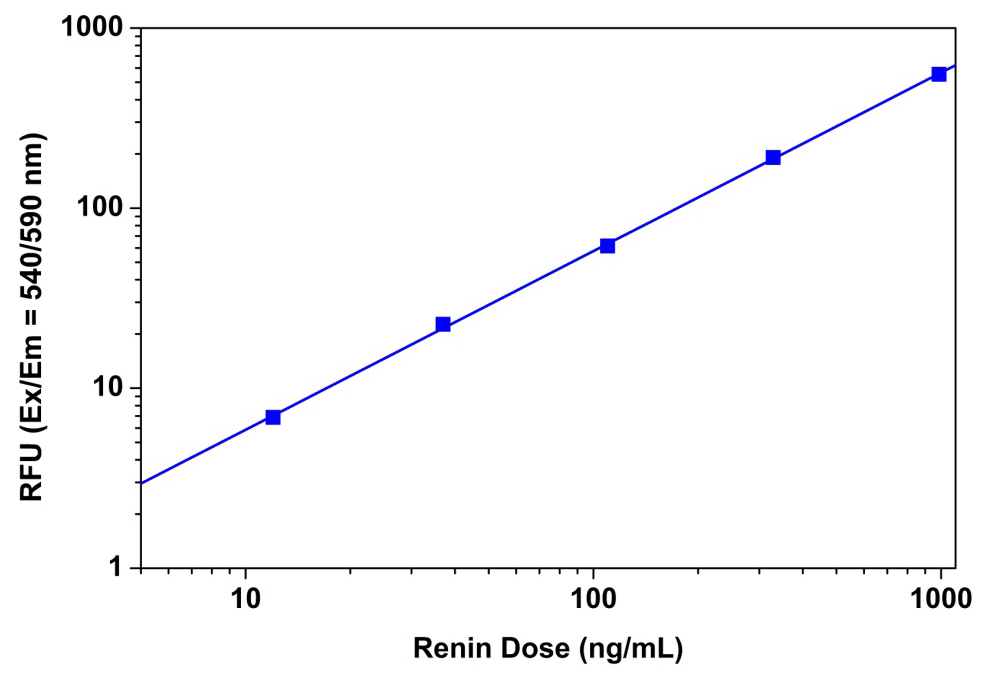
Figure 4. Renin dose responses were measured with Amplite™ Fluorimetric Renin Assay Kit on a 96-well black solid plate using a Gemini fluorescence microplate reader (Molecular Devices). As low as 0.01 µg/mL renin was detected with 30 minutes incubation (n=3).
Peptidases
Peptidases and proteases play essential roles in protein activation, cell regulation and signaling, as well as in the generation of amino acids for protein synthesis or utilization in other metabolic pathways. In general, peptidases cleave shorter peptides, and proteases cleave longer peptides and proteins. Depending on their sites of cleavage, peptidases can be classified as exopeptidases if they preferentially hydrolyze amino acid residues from the terminus of a peptide, or endopeptidases if they cleave internal peptide bonds. Exopeptidases are further divided into aminopeptidases and carboxypeptidases depending on whether they hydrolyze residues from the amine or the carboxy terminus.
Although the spectral properties of fluorogenic peptidase and protease substrates and their hydrolysis products are easily predictable, the utility of a given substrate for an enzyme depends on the kinetics of hydrolysis by the enzyme, which, in turn, depends on the substrate’s concentration and amino acid sequence, as well as on the pH, temperature and presence of cofactors in the medium.
7-Amino-4-methylcoumarin (AMC)
7-Amino-4-methylcoumarin (AMC) is a blue-fluorescent dye whose peptide amides are used extensively as substrates for detecting enzymatic activity in cells, homogenates and solutions. 7-Amino4-trifluoromethylcoumarin (AFC) is a dye with somewhat longer wavelength spectra than AMC (excitation/emission maxima of ~380/500 nm) at pH 7. AAT Bioquest offers a variety of substrates based on AMC and AFC.
Rhodamine 110 (R110)
Rhodamine 110 (R110) is a visible light–excitable dye with much stronger absorption than AMC. R110-based substrates usually comprise two identical amino acids or peptides attached to a single fluorophore. AAT Bioquest’s bisamide derivatives of rhodamine 110 are sensitive and selective substrates for assaying protease activity in solution or inside live cells. These substrates comprise an amino acid or peptide covalently linked to each of R110’s amino groups, thereby suppressing both its visible absorption and fluorescence. Upon enzymatic cleavage, the nonfluorescent bisamide substrate is converted in a two-step process first to the fluorescent monoamide and then to the even more fluorescent R110. The fluorescence intensities of the monoamide and of R110 are constant from pH 3–9. Both of these hydrolysis products exhibit spectral properties similar to those of fluorescein, with peak excitation and emission wavelengths of 496 nm and 520 nm, respectively, making them compatible with flow cytometers and other instrumentation based on the argon-ion laser. Substrates based on R110 may also be useful for sensitive spectrophotometric assays because the R110 dye has intense visible absorption (extinction coefficient at 496 nm =?80,000 cm-1M-1 in pH 6 solution).
| Product Name | Mol. Wt. | Ex (nm) | Em (nm) | Filter Set | Protease |
| Ala-AMC | 360.28 | 339 | 439 | DAPI | Alanyl aminopeptidase and trypsin |
| BOC-Val-Pro-Arg-AMC | 741.75 | 339 | 439 | DAPI | Alpha-thrombin |
| (BOC-VPR)2R110 | 1463.48 | 499 | 521 | FITC | Alpha-thrombin |
| D-Ala-AMC *CAS: 201847-52-1* | 360.28 | 339 | 439 | DAPI | Aminopeptidase and endopeptidase |
| D-VLK-AMC [D-Val-Leu-Lys-AMC] | 743.69 | 339 | 439 | DAPI | Plasmin |
| Gly-Pro-AMC *CAS: 115035-46-6* | 443.37 | 339 | 439 | DAPI | Dipeptidyl peptidase IV (DPPIV) |
| MeO-Succ-Arg-Pro-Tyr-AMC | 819.78 | 339 | 439 | DAPI | Chymotrypsin-like proteases |
| Pro-AMC | 272.3 | 339 | 439 | DAPI | Tricorn Protease (TRI) |
| Benzoyl-Arg-AMC | 549.5 | 339 | 439 | DAPI | Human alpha-thrombin |
| Z-Gly-Pro-AMC *CAS: 68542-93-8* | 463.49 | 339 | 439 | DAPI | Post-proline cleaving enzyme (prolyl endopeptidase) |
| Benzoyl-Leu-Gly-Arg-Aminoluciferin | 823.86 | N/A | 560 | N/A | Luminogenic substrate for complement component C3/C5 convertases, coagulation factor Xa, macropain, and soybean trypsin-like enzyme |
Table 6. Ordering Info for Generic Protease Probes and Assays Products
Table 7. Ordering Info for MMP Probes and Assays Products
| Cat# | Product Name | Unit Size |
| 13510 | Amplite™ Universal Fluorimetric MMP Activity Assay Kit *Green Fluorescence* | 100 Tests |
| 13511 | Amplite™ Universal Fluorimetric MMP Activity Assay Kit *Red Fluorescence* | 100 Tests |
| 13512 | Amplite™ MMP-3 Activity Assay Kit *Green Fluorescence* | 100 Tests |
| 13519 | MMP Green™ substrate | 100 Tests |
| 13520 | MMP Green™ substrate | 1 mg |
| 13521 | MMP Red™ substrate | 1 mg |
| 13522 | MMP Red™ substrate | 100 Tests |
| 13527 | MMP-3 Green™ substrate | 100 Tests |
| 13528 | MMP-3 Green™ substrate | 1 mg |
Table 8. Ordering Info for Proteasome 20S Probes and Assays Products
| Cat# | Product Name | Unit Size |
| 13451 | (Suc-LLVY)2R110 | 1 mg |
| 13452 | Suc-LLVY-Aminoluciferin | 1 mg |
| 13453 | Suc-LLVY-AMC | 1 mg |
| 13455 | (Ac-ANW)2R110 | 1 mg |
| 13456 | Amplite™ Fluorimetric Proteasome 20S Activity Assay Kit *Green Fluorescence* | 100 tests |
| 13465 | (Ac-KQL)2R110 | 1 mg |
| 13466 | (Z-LLE)2R110 | 1 mg |
| 13467 | (Ac-PAL)2R110 | 1 mg |
| 13468 | (Ac-WLA)2R110 | 1 mg |
Table 9. Ordering Info for Renin Probes and Assays Products
| Cat# | Product Name | Unit Size |
| 13530 | Amplite™ Fluorimetric Renin Assay Kit *Red Fluorescence* | 100 tests |
Table 10. Ordering Info for Peptidase Probes and Assays Products
| Cat# | Product Name | Unit Size |
| 13000 | Benzoyl-Leu-Gly-Arg-Aminoluciferin | 1 mg |
| 13201 | D-VLK-AMC [D-Val-Leu-Lys-AMC] | 5 mg |
| 13450 | Gly-Pro-AMC *CAS 115035-46-6* | 5 mg |
| 13457 | Pro-AMC | 5 mg |
| 13458 | Ala-AMC | 5 mg |
| 13459 | D-Ala-AMC *CAS 201847-52-1* | 5 mg |
| 13460 | MeO-Succ-Arg-Pro-Tyr-AMC | 5 mg |
| 13461 | BOC-Val-Pro-Arg-AMC | 5 mg |
| 13462 | (BOC-VPR)2R110 | 1 mg |
| 13477 | Z-Gly-Pro-AMC *CAS 68542-93-8* | 5 mg |
| 13478 | Benzoyl-Arg-AMC | 5 mg |