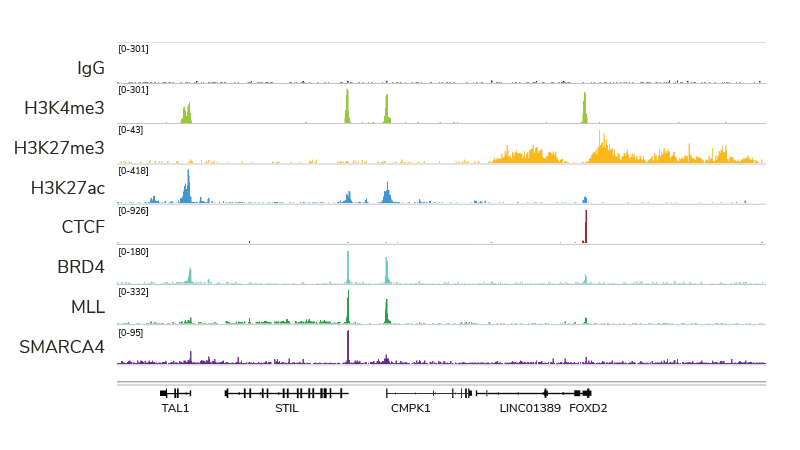
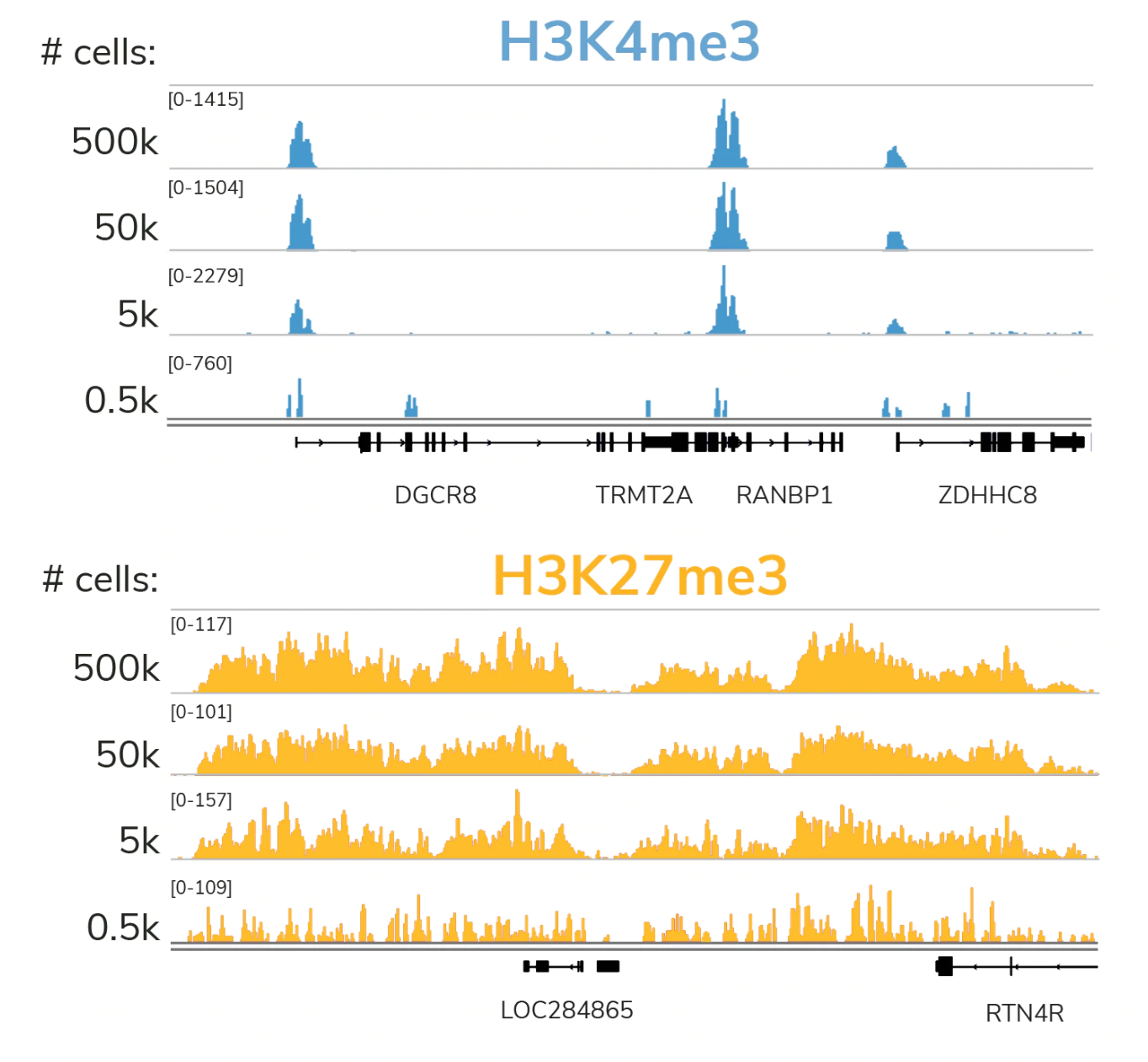
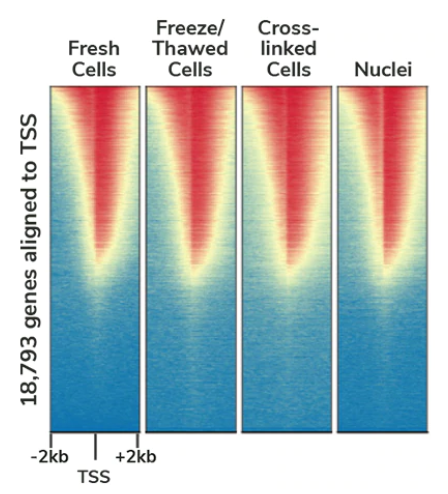
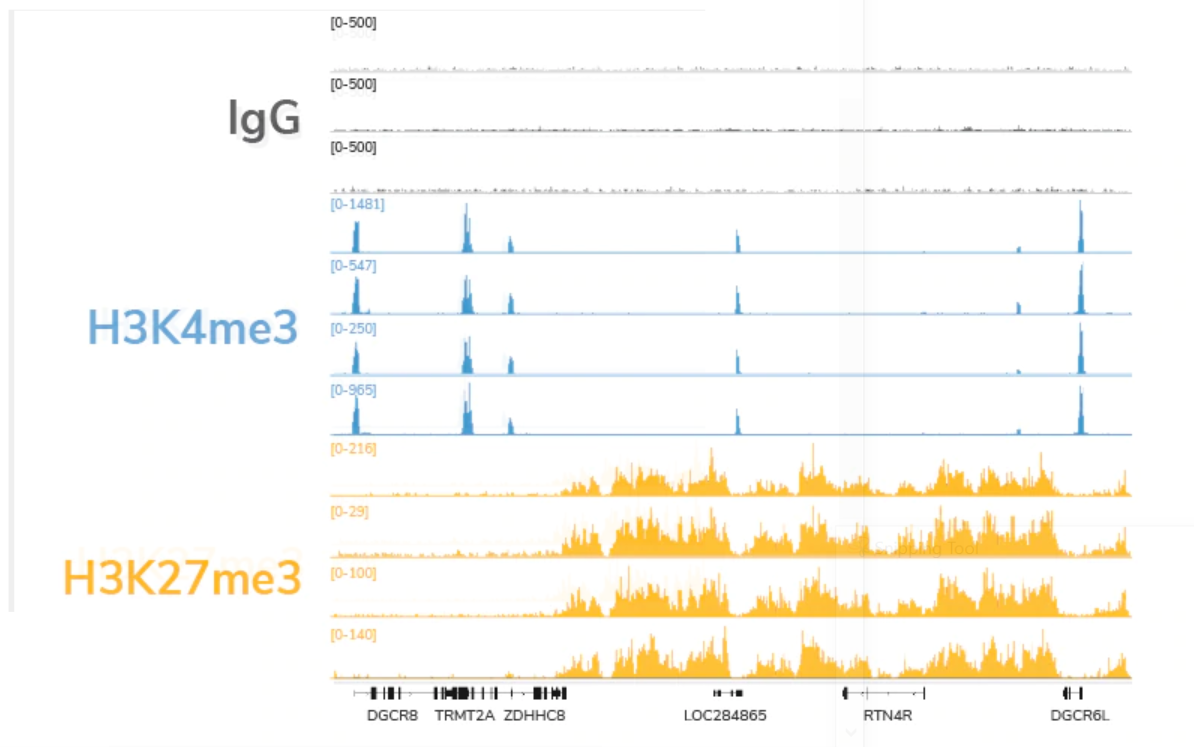
Cleavage Under Targets and Release using Nuclease (CUT&RUN), based on the previous Chromatin ImmunoCleavage (ChIC) approach, uses a fusion of protein A, protein G, and micrococcal nuclease (pAG-MNase) to cleave and extract antibody-bound chromatin. The targeted nature of CUT&RUN’s pAG-MNase improves signal : noise and reduces sequencing costs resulting in a highly cost- and time-efficient NGS assay.
Since launching pAG-MNase, EpiCypher has spent the last year launching a series of CUT&RUN companion products, meticulously hand-selected based on top CUT&RUN performance. Our kit is the culmination of these efforts, providing customers with a standout tool for accurate and user-friendly chromatin profiling.
- Diverse target compatibility
- Low cell input requirements
- Detailed manual with common sample preparation methods
- Highly robust and reproducible workflow
- Access to exclusive quality controls