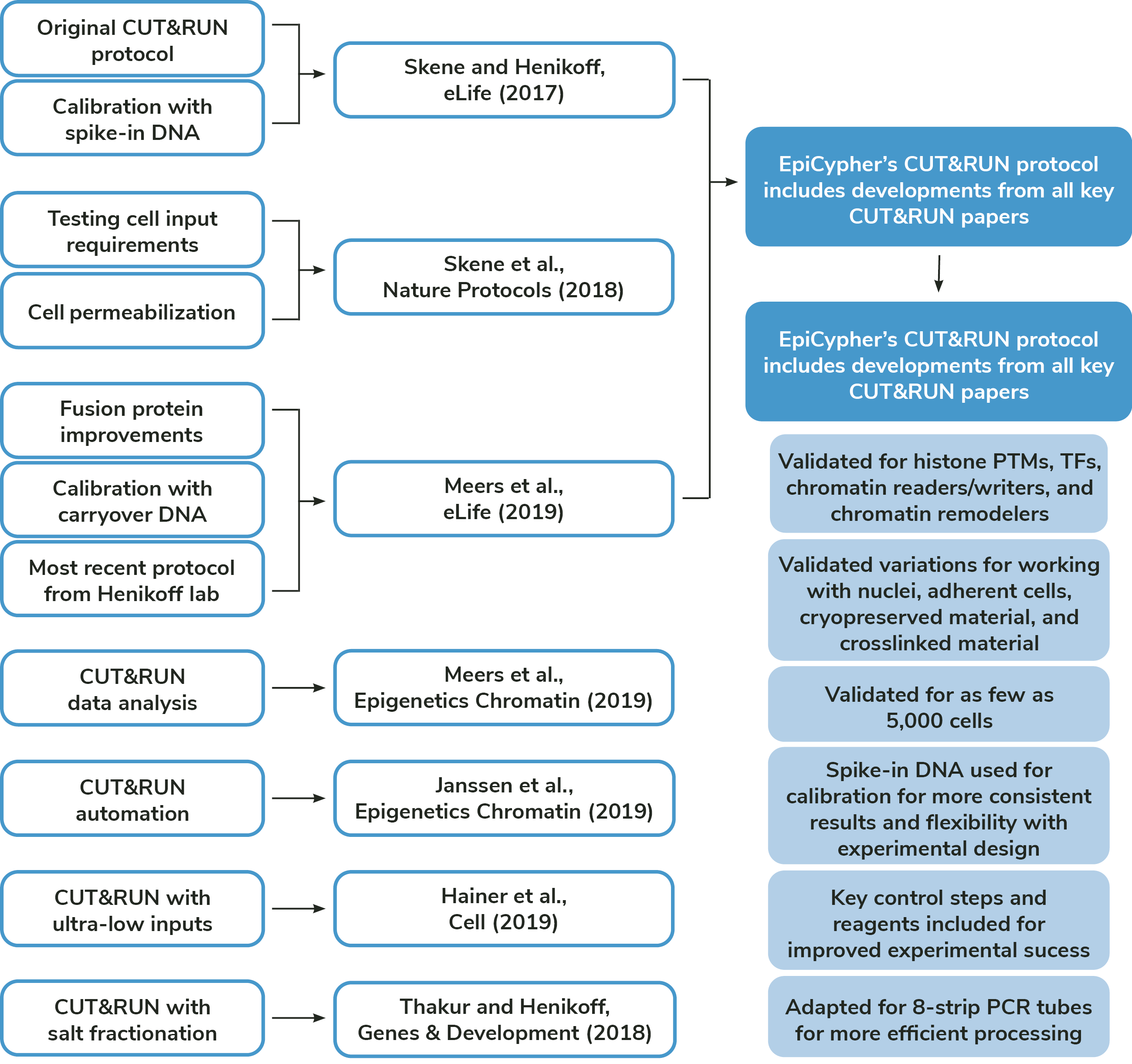
Foundational CUT&RUN publications
An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites
Authors: Skene and Henikoff
Published: eLife, 2017
PMID: 28079019
Key Takeaways:
- First publication describing CUT&RUN as a novel method for chromatin profiling, an evolution of chromatin immunocleavage (ChIC)
- Protein A-MNase (pA-MNase) used to cleave antibody-bound loci in intact native nuclei, from as few as 600,000 cells
- Nuclei immobilized on magnetic Concanavalin A (ConA) beads to streamline sample processing, allowing CUT&RUN assay to be completed in one day
- Applied CUT&RUN to map transcription factors (TFs), histones, and chromatin regulators
- Utilized Drosophila spike-in DNA for normalization
- Confirmed that CUT&RUN performs as well as or better than ChIP-seq with respect to simplicity, resolution, robustness, efficiency, data quality, and applicability to highly insoluble complexes
Targeted in situ genome-wide profiling with high efficiency for low cell numbers
Authors: Skene, Henikoff, and Henikoff
Published: Nature Protocols, 2018
PMID: 29651053
Key Takeaways:
- Introduced improvements to the original CUT&RUN protocol
- Eliminated need to isolate nuclei by introducing cell permeabilization with digitonin, which permeates cell membranes without compromising nuclear integrity
- Demonstrated drastically reduced cell input requirements by generating high-quality data from as few as 100 cells for an abundant histone PTM or 1,000 cells for a TF
Improved CUT&RUN chromatin profiling tools
Authors: Meers, Bryson, Henikoff, and Henikoff
Published: eLife, 2019
PMID: 31232687
Key Takeaways:
- Introduced further improvements to the CUT&RUN protocol
- Updated nuclease fusion protein construct from pA-MNase to a pAG-MNase (hybrid protein A-protein G-MNase)
- Expands compatibility to mouse antibodies
- Mutations in protein G increase binding to rabbit antibodies
- Added histidine tag to the construct to improve purification
- Modified MNase digestion protocol under high-calcium/low-salt conditions
- Inhibits premature release of the nuclease-bound complex
- Reduces risk of over-digestion for abundant epitopes
- Described calibration strategy without use of spike-in
- Leverages carry-over E. coli DNA introduced by addition of MNase fusion protein (purified from E. coli)
- Note: EpiCypher pAG-MNase preps lack sufficient carry-over DNA for calibration; we offer spike-in E. coli DNA for normalization, and CUT&RUN-compatible dNuc spike-in controls are in development
Peak calling by Sparse Enrichment Analysis for CUT&RUN chromatin profiling
Authors: Meers, Tenenbaum, and Henikoff
Published: Epigenetics Chromatin, 2019
PMID: 31300027
Key Takeaways:
- Described SEACR (Sparse Enrichment Analysis for CUT&RUN), a method for peak calling that is compatible with low-background CUT&RUN data
- Overcomes false-positive peak calling problems in CUT&RUN data analysis, caused by using algorithms originally designed for analysis of lower-sensitivity ChIP-seq
Advanced CUT&RUN publications
For users interested in further documented development of the CUT&RUN workflow for more advanced and/or specific applications, below we highlight three additional publications that are expanding the capabilities of this innovative methodology.
Automated in situ chromatin profiling efficiently resolves cell types and gene regulatory programs
Authors: Janssens, Wu, Sarthy, Meers, Myers, Olson, Ahmad, and Henikoff
Published: Epigenetics Chromatin, 2018
PMID: 30577869
Key Takeaways:
- Modified CUT&RUN protocol for a 96-well plate format on a liquid handling robot (autoCUT&RUN)
- Applied autoCUT&RUN protocol to profile cells and frozen tumor samples
- Identified cell-type specific promoter and enhancer activities, illustrating power of platform
Profiling of Pluripotency Factors in Single Cells and Early Embryos
Authors: Hainer, Boskovic, McCannell, Rando, and Fazzio
Published: Cell, 2019
PMID: 30955888
Key Takeaways:
- Adapted the CUT&RUN method for ultra-low inputs (uliCUT&RUN)
- Modified multiple protocol steps including alteration of buffers, sample volumes, incubation times, quantities of spike-in DNA, and methods for library preparation and purification
- Applied uliCUT&RUN to profile both a PTM and a TF using 10 cells per target
- Demonstrated specific (albeit sparse) TF profiling in single cells
- Further adapted uliCUT&RUN protocol to profile TFs in pre-implantation embryos
Unexpected conformational variations of the human centromeric chromatin complex
Authors: Thakur and Henikoff
Published: Genes Dev, 2018
PMID: 29386331
Key Takeaways:
- Combined CUT&RUN with salt fractionation to map chromatin-associated proteins from both soluble and insoluble chromatin (CUT&RUN.Salt)
- Applied CUT&RUN.Salt to profile specific centromeric chromatin complexes (e.g. CENP-A), which is insoluble under conditions typically used for native chromatin extraction
Excited about the potential of CUT&RUN? You’re not alone! EpiCypher is working hard to stay on the cutting edge of the development and application of this transformative method for chromatin profiling. Our CUTANA™ CUT&RUN kit and optimized protocols make it easier than ever to bring CUT&RUN into your laboratory and leave expensive, time-and resource-intensive ChIP-seq methods in the past.