New automatised tools are required to process high-throughput time-lapse imaging data of bacterial cells. DeLTA (Deep Learning for Time-lapse Analysis) is a rapid and accurate image analysis workflow that uses convolutional neural networks (U-Net) to segment and track single cells growing on 2D surfaces. It is available open source and uses a fully Python implementation [1].
An open-source deep-learning tool for segmenting and tracking bacteria
Figure 1: Segmentation and Tracking in 2D [https://doi.org/10.1371/journal.pcbi.1009797, PLOS]
Deep learning is necessary to automatise single-cell data analysis in large datasets
Traditional microbiology usually studies microbes at population-level, without considering the individuals’ heterogeneity in a population. The study of this heterogeneity is essential to understand how individuals respond to environmental changes and determine the fate of the whole population [2].
For this reason, single-cell microfluidic experiments are leading important discoveries in different fields, such as cell-size control, cellular aging, stress responses and antibiotic tolerance/ persistence.
A direct consequence is that improvements in time-lapse microscopy are significantly increasing the size of the new datasets. Traditional approaches require time-consuming manual error correction, and image analysis is becoming a major bottleneck in generating quantitative, single-cell data.
In recent years, deep learning tools have found applications in many biology fields, including automated image analysis [3]. For example, the U-Net algorithm is a convolutional neural network that performs exceptionally well on biomedical data [4].
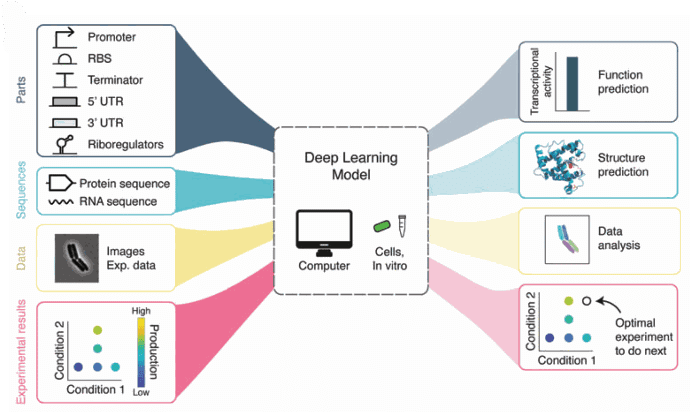
Figure 2: The applications of Deep learning in biology. We are interested in image analysis. [ https://doi.org/10.1089/genbio.2022.0017, GEN Biotech]
A purely Python workflow with two U-Net convolutional neural networks
An ideal tool for high throughput analysis of microscopy data has to be rapid, accurate, flexible and require minimal input from the user. DeLTA addresses this combination of needs thanks to a deep-learning-based approach.
In addition, the purely Python workflow allows the entire pipeline to be in an open-source framework. The Bio-Formats toolbox for Python also enables users to work directly with images in many standard formats without needing any preformatting.
To summarise, DeLTA pipeline consists of segmentation, tracking, and lineage reconstruction.
Both the segmentation and tracking models implement a U-Net neural network architecture (TensorFlow/Keras). A pixel-wise-weighted binary cross-entropy loss function is implemented to train both models, as in the original U-Net paper [4].
As input for segmentation, images in many common formats that are output via microscopy software can be used without the need for any preformatting. The tracking model inputs a phase contrast image and segmentation from the current and previous frame and outputs a greyscale image of the predicted tracking event.
Ad hoc functions are used to deal with occasional out-of-focus frames, electronic noise, exaggerated cell movement and drift.
As a result, segmentation generates information about cell morphology; tracking tracks cells across frames and records division events; lineage reconstruction keeps track of cell lineages and records pole age.
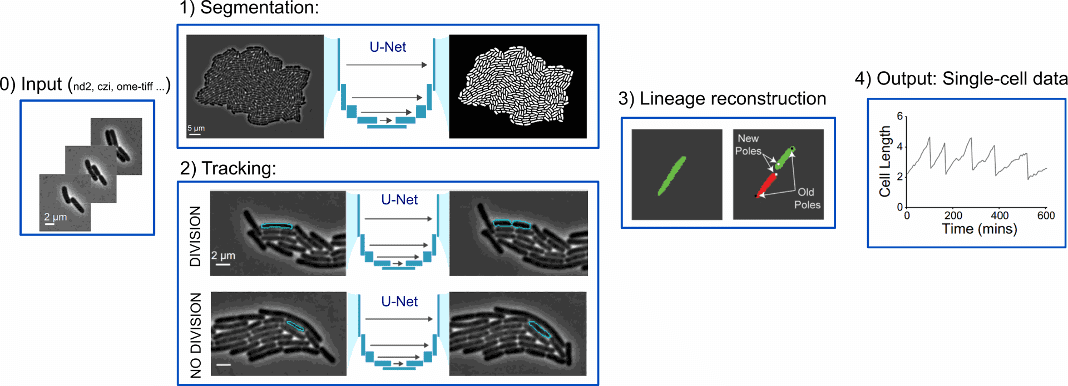
Figure 3: DeLTA pipeline [ Adapted from https://doi.org/10.1371/journal.pcbi.1009797, PLOS]
New functionality to analyse mixed populations in two dimensions
While the first version of DeLTA was working in 1D [5], this new version segments and tracks bacteria in 2D. Moreover, cells with different properties can be tracked simultaneously within the same movie.
All this is essential to quantify single-cell spatial and temporal dynamics, which we know to be necessary to study microbial co-cultures, cell-to-cell interactions, or spatial patterning.
A possible application is to look at differences in growth rate between antibiotic-resistant and susceptible cells grown in the same field of view.
DeLTA works very well on standard rod-shaped bacteria such as E. coli and B. subtilis. It can be easily extended to cells with similar morphologies, but also to different organisms.
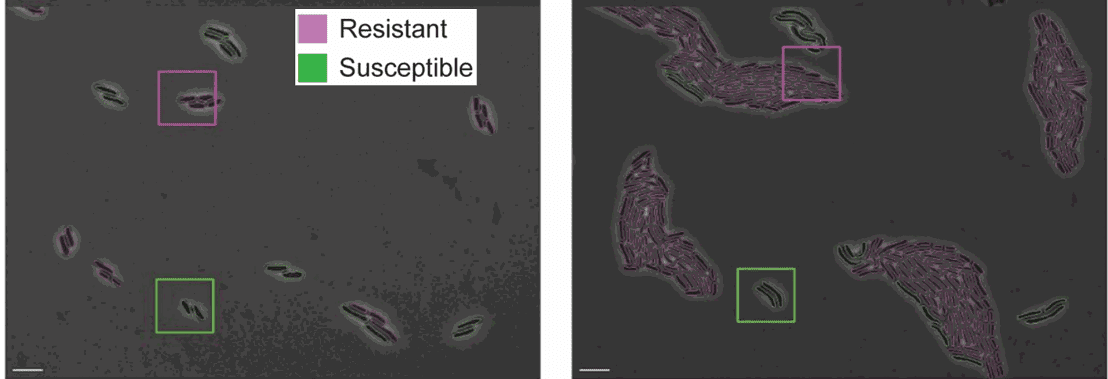
Figure 4: Resistant and susceptible strains of E. coli on agarose pads containing an inhibitory concentration of tetracycline [https://doi.org/10.1371/journal.pcbi.1009797, PLOS]
Why did I like this tool?
DeLTA is a powerful tool for analysing 2D time-lapse microscopy data. It is an entirely open-source Python workflow, and it accepts many standard microscopy file formats as inputs.
Moreover, a Google Colab notebook is available to test it with your own data without installing the code on your local machine [6]. You have no excuse not to try it!
References:
[1] https://delta.readthedocs.io/en/latest/
[2] Allard P, Papazotos F, Potvin-Trottier L. Microfluidics for long-term single-cell time-lapse microscopy: Advances and applications. Front Bioeng Biotechnol. (2022) https://doi.org/10.3389/fbioe.2022.968342
[3] Beardall WAV, Stan GB, Dunlop MJ. Deep Learning Concepts and Applications for Synthetic Biology. GEN Biotechnol. (2022) https://doi.org/10.1089/genbio.2022.0017
[4] Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. ArXiv (2015) https://doi.org/10.48550/arXiv.1505.04597
[5] Lugagne J-B, Lin H, Dunlop MJ. DeLTA: Automated cell segmentation, tracking, and lineage reconstruction using deep learning. PLoS Comput Biol (2020) https://doi.org/10.1371/journal.pcbi.1007673
[6] https://colab.research.google.com/drive/1UL9oXmcJFRBAm0BMQy_DMKg4VHYGgtxZ.
