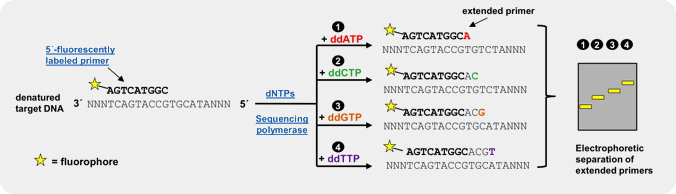
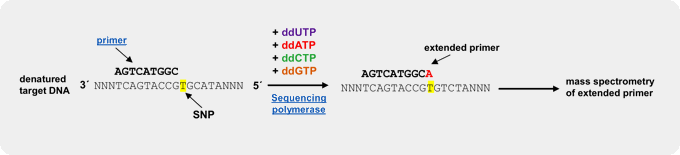
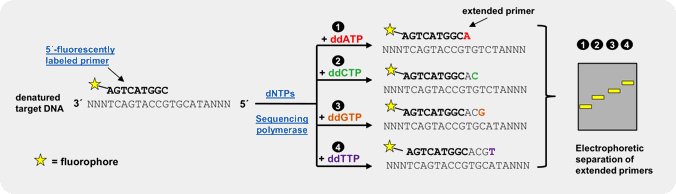
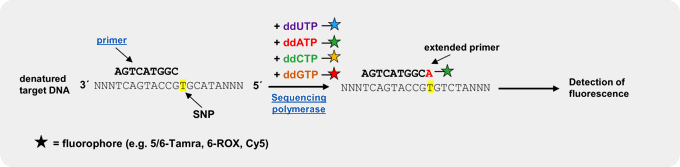
Fluorescently labeled dideoxy-Nucleoside triphosphates (ddNTPs)
Single nucleotide polymorphisms (SNPs) are single base pair mutations at specific locations in the coding and non-coding regions of the genome that are found in more than 1% of a population[1]. While SNPs located within a coding region are most likely to alter the biological function of a protein, SNPs found within the non-coding regions may affect gene regulation. Both, coding and non-coding SNPs have been associated with population-specific disease development or drug susceptibility and are therefore promising molecular markers for disease genetics and pharmacogenomics studies[2].
An efficient and reliable approach for genotyping of a priori known SNP locations is single basepair extension (SBE)[3,4,5]. This method relies on the extension of a primer, designed to bind one nucleotide upstream of the polymorphic spot, by a SNP-complementary, fluorescently labeled ddNTP[5]. Subsequent detection of the incorporated ddNTP is performed by fluorescent visualization of the extended primer by electrophoresis or array-based methods, thus revealing the nucleotide base at that position on the template strand.
Our fluorescently labeled ddNTPs are > 95 % pure (HPLC) and have been successfully used in SBE experiments[5]. Further fluorescently labeled ddATP, ddCTP, ddGTP and ddUTP analogs (e.g. Cy3 or Cy5-labeled), amine-labeled ddNTPs for subsequent coupling of NHS-ester-modified fluorescent dyes → Probes & Epigenetics as well as biotin-labeled ddNTPs for subsequent detection with fluorescent streptavidin are available as catalog products.
Bulk Amounts
If you require large amounts (> 10 ml, > 100 ml, > 1 l), significant discounts are available. Please contact us at info@stratech.co.uk to receive an individual quotation.
Selected References
[1] Brookes et al. (1999) The essence of SNPs. Gene 234:177.
[2] Kim et al. (2007) SNP genotyping: technologies and biomedical applications. Annual review of Biomedical Engineering 9:289.
[3] Desphande et al. (2005) Multiplexed SNP Genotyping using single-base extension (SBE) and microsphere arrays. Current protocols in Cytometry 13 (4):1.
[4] Syvänen et al. (1999) From gels to chips: Minisequencing primer extension for analysis of point mutations and single nucleotide polymorphisms. Hum. Mut. 13:10.
[5] Esteves et al. (2011) Clinical relevance of multiple single-nucleotide polymorphisms in Pneumocystis jirovecii Pneumonia: development of a multiplex PCR-single-base-extension methodology. J. Clin. Microbiol. 49 (5):1810.