Antibody Selection
The most critical step in the development of an ELISA is the selection of antibodies, and the criterium upon which each antibody is selected is based upon its function within the particular ELISA format selected.
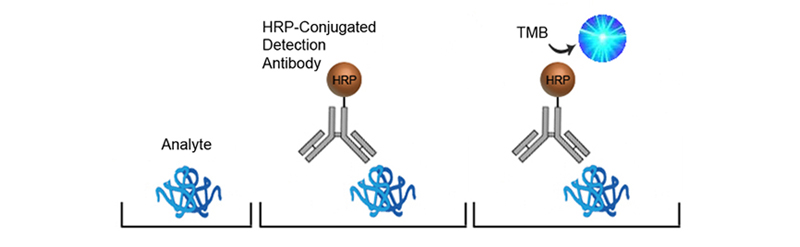
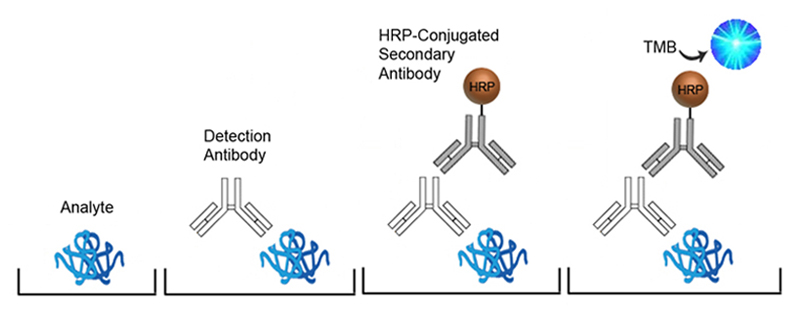
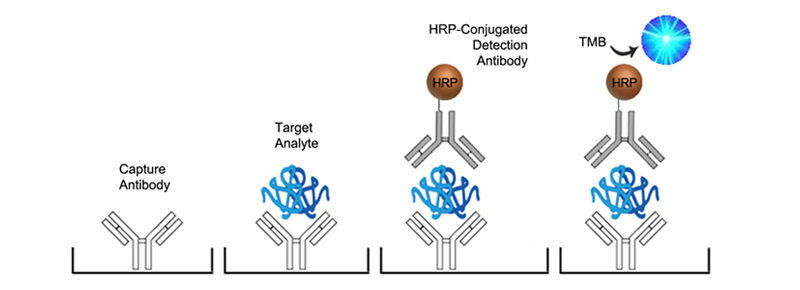
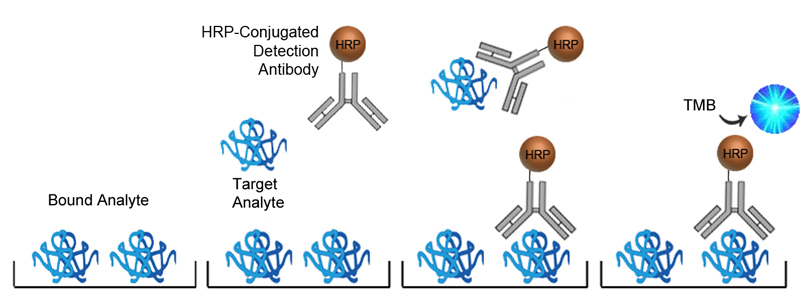
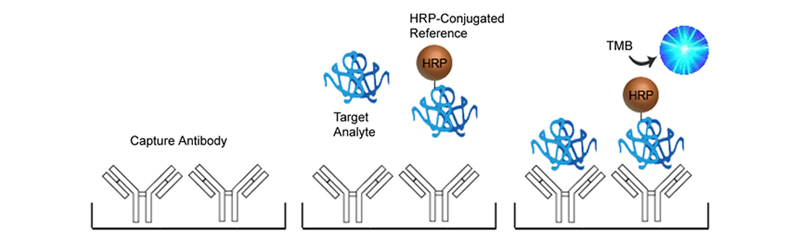
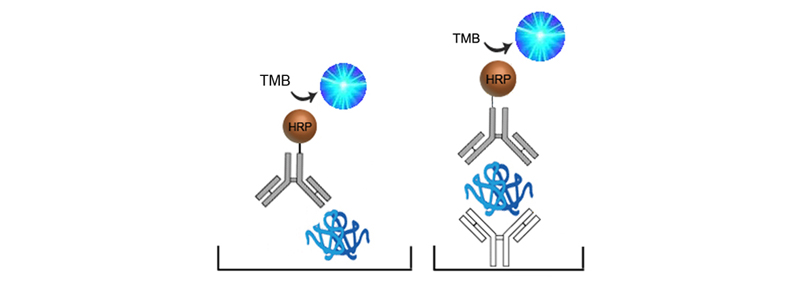
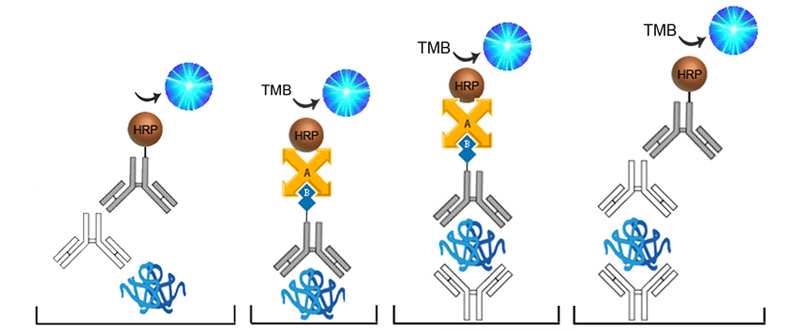
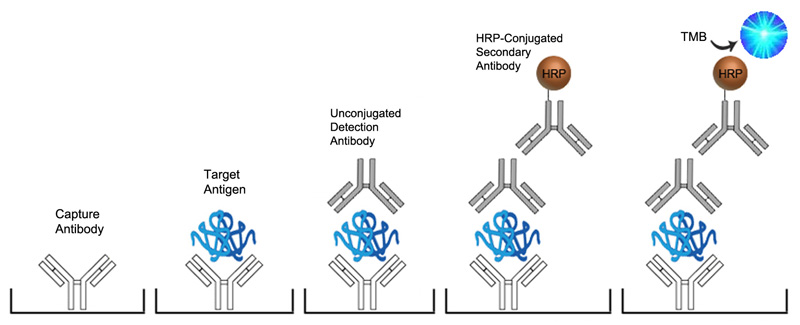
In Sandwich ELISAs for example, a capture and detection antibody are needed. These antibodies can be either monoclonal or polyclonal but the pros and cons of each type should be considered. Monoclonal antibodies are highly specific for a single unique epitope on their target antigen. For this reason they cannot be used as both the capture and detection antibody in the same assay as they would compete for the same binding site. This highly selective binding characteristic can make monoclonal antibodies highly specific, but potentially at the cost of sensitivity as fewer binding sites are availabl on the target analyte. Monoclonals are also highly reproducible, making them ideal for developing assays intended for long term production. In contrast, each polyclonal antibody is a heterogeneous collection of antibodies, targeting multiple binding sites on the target analyte, from stable distinct peptide regions to tertiary structural epitopes that can change with assay conditions. For this reason, polyclonal antibodies have the potential to be less specific than monoclonals, but the fact that they target multiple epitopes make them more flexible in their use, and less sensitive to changing assay conditions.
The specificity of each antibody used in an ELISA also depends upon its intended role. For example, in a Sandwich ELISA, a more specific monoclonal antibody may be used as the capture antibody to ensure that only the target antigen is captured, while a less discriminating polyclonal antibody is used for detection ensuring maximum signal detection.
Antibody Conjugation
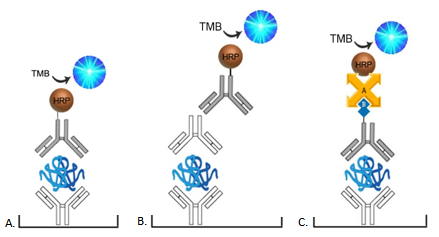
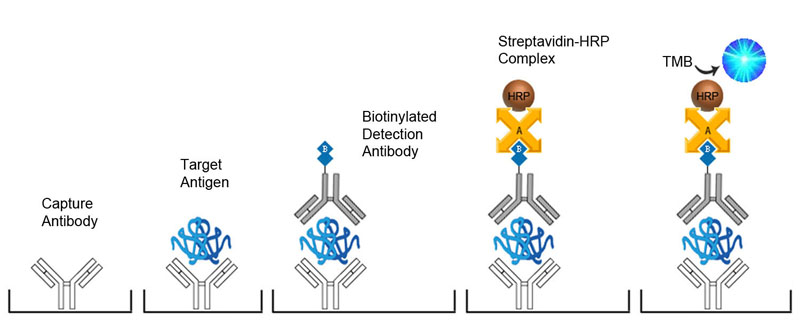
Depending upon the type of ELISA format selected, and the availability of antibodies, it may be necessary to conjugate antibodies directly with an enzyme, such as Horseradish Peroxidase (HRP) or alkaline phosphatase (AP), or with Biotin. Contact us at technical@stratech.co.uk to inquire about recommended conjugation kits.
Antibody Titration
Once an antibody pair has been selected, it is recommended to determine the optimal concentration of each by running a dilution matrix assay.
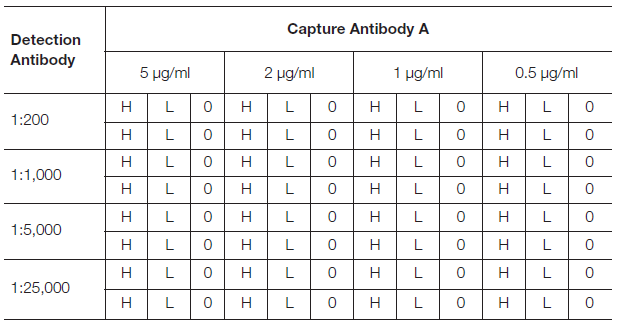
Prepare 4-step dilutions of the capture antibody in coating buffer (5, 2, 1, and 0.5 μg/ml) and coat the wells of a 96-well plate as per Table 1. Next, follow the standard procedure for a Sandwich ELISA using high and low concentrations of your analyte that reflect the expected working range, and a blank. At the detection stage, prepare a dilution series of the detection antibody in dilution buffer (1:200, 1:1,000, 1:5,000, and 1:25,000) and apply as per Table 1.
Including a high and low concentration of the analyte helps to determine the dynamic range. The low concentration analyte indicates the sensitivity of the assay. The blank will indicate non-specific binding. The optimal combination will have the maximum signal-to-noise ratio/largest difference between low and high analyte concentrations.
If the blank samples indicate high background, readings above 0.2 absorbance units, consider changing the ELISA plate type, the blocking buffer, and/or the washing buffers used. If sensitivity is not high enough, consider modifying the sample buffer to limit matrix effect, wash conditions, and incubation times.