Multiple labeling is the process of sequential immunolabeling to detect multiple antigens by either immunofluorescent or colorimetric IHC/ICC. The successful detection of more than one antigen requires rational experimental design, taking into account a large number of variables accumulated by each of the experimental steps. Here we detail how to set up the multiple labeling experiment and some considerations essential to executing a successful assay.
Multiple labeling for simultaneous detection of several targets
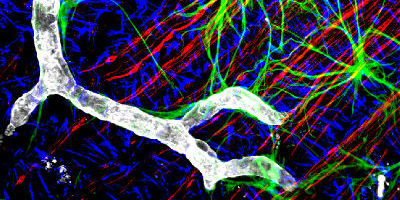
Mouse GFAP (green), NF (red), Collagen IV (grey), Vimentin (blue) z1. Image courtesy of Gabe Luna, Neuroscience Research Institute, UC Santa Barbara.
Designing the multiple labeling experiment
Selection of antibodies for simultaneous detection of more than one antigen depends on at least two important criteria:
- Availability of secondary antibodies that do not recognize:
- One another (are derived from the same host species),
- Other primary antibodies used in the assay system,
- Endogenous immunoglobulins present in the tissues or cells under investigation.
- Use of probes (enzyme-reaction products, fluorophores, or electron-dense particles) that are well resolved.
The affinity-purified antibodies marked “ML” (multiple labeling) have been specifically prepared to meet these criteria. They include mouse IgG subclass specific antibodies and antibodies cross-adsorbed (min X) against other species.
Multiple Labeling Example
| Sample | Mouse | Mouse | Mouse |
| Antigen | Antigen A | Antigen B | Antigen C |
| Blocking Step | Step 1 | Step 4 | Step 7 |
| 5% Normal Donkey serum to block | 5% Normal Donkey serum to block (if needed) | 5% Normal Donkey serum to block (if needed) | |
| Wash | Wash | Wash | |
| Primary Antibody Step | Step 2 | Step 5 | Step 8 |
| Goat Anti-Antigen A | Rabbit Anti-Antigen B | Rat Anti-Antigen C | |
| Wash | Wash | Wash | |
| Secondary Antibody Step | Step 3 | Step 6 | Step 9 |
| Probe 1 Donkey Anti-Goat IgG (H+L) (min X Ck, GP, Sy Hms, Hrs, Hu, Ms, Rb, Rat Sr Prot) | Probe 2 Donkey Anti-Rabbit IgG (H+L) (min X Bov, Ck, Gt, GP, Sy Hms, Hrs, Hu, Ms, Rat, Shp Sr Prot) | Probe 3 Donkey Anti-Rat IgG (H+L) (min X Bov, Ck, Gt, GP, Sy Hms, Hrs, Hrs, Hu, Ms, Rb, Shp Sr Prot) |
Note: In this example, the secondary antibodies used do not recongnize each other since they are all made in donkey. They have been solid-phase adsorbed so that they do not recognize the other primary antibodies used (min X) in steps, 2, 5, and 8. Also, they do not react with endogenous mouse Ig, which may be present in mouse tissue.
Caution: Do not dilute any antibody with normal serum or mix antibodies together to save time, as this may result in immune complex formation and increased background.
Labeling primary antibodies from the same host species
Monovalent Fab fragments of affinity-purified secondary antibodies are offered to cover (block) the surface of immunoglobulins for double labeling primary antibodies from the same host species, or to block endogenous immunoglobulins in tissue sections or on cell surfaces. They can be used for these purposes because Fab fragments have only a single antigen binding site (i.e. they are monovalent).
Labeling primary antibodies from the same species as the sample
Sometimes it may be necessary to use a primary antibody from the same species as the sample, for example Mouse on Mouse. In these circumstances secondary antibodies may also detect endogenous immunoglobulin, causing unwanted background. This can be avoided with Fab fragment blocking.
Further Considerations
Blocking
For general blocking purposes, normal serum (5% v/v) from the same species as the secondary antibody host provides efficient background reduction for non-specific, conserved-sequence, and/or Fc-receptor binding.
Specific unwanted reactions with antibodies can be blocked with monovalent Fab fragments of secondary antibodies. This type of blocking is indicated for situations in which the specimen and primary antibodies are of the same species (e.g.mouse on mouse labeling), or when multiple primary antibodies are raised in the same host animal.
Visualization
Successful multiple labeling depends on the use of probes whose signals can be distinguished by available equipment.
Fluorescence microscopy is a common platform for multiple labeling, since filter sets have been designed to discriminate among the many fluorophores available. Narrow band-pass emission filters are critical for separating signals from multiple fluorophores, suppressing detection of fluorescence from overlapping spectra. When planning a multiple labeling protocol, formulate a dye panel from fluorophores with well separated emission spectra that are compatible with available instrumentation.
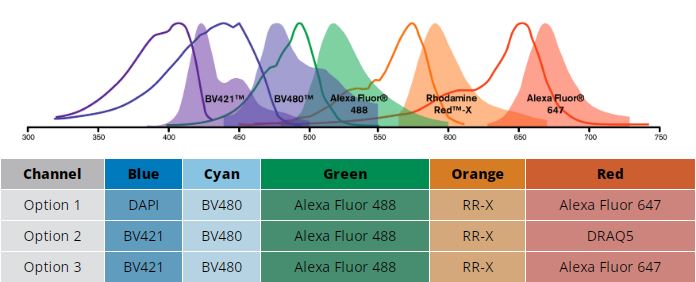
Three examples of dye panels. Options 1 and 2 combine nuclear stains with immunofluorescence. Option 3 shows 5 color immunofluorescence.
Multiple labeling can also be achieved with enzyme-linked antibodies. An antigenic site is labeled with a primary and secondary antibody, followed by color development with a chromogenic substrate such as DAB, TMB or AEC. Additional antigenic sites are labeled sequentially, with different chromogens used for each antigenic site.
For multiple labeling in electron microscopy, different sizes of colloidal gold particles complexed with secondary antibodies allow clear visualization of separate antigenic sites.
Controls
Prior to performing a multiple labeling protocol, optimize conditions for each primary/secondary antibody pair. Titrating both the primary and secondary antibody will identify conditions with low background and best positive signal. To demonstrate the specificity of each secondary antibody for its intended primary, attempt to label primaries with the “wrong” secondary antibodies (negative controls).
When using primary antibodies raised in the same host as the specimen species (e.g. mouse on mouse), controls can be used to determine the level of non-specific signal.
For a review of multi-color immunofluorescence labeling with confocal microscopy see Brelje, Wessendorf, and Sorenson, “Multi-color laser scanning confocal immunofluorescence microscopy: Practical application and limitations.” In Cell Biological Applications of Confocal Microscopy (Methods in Cell Biology. vol. 38). Ed. B. Matsumoto. Orlando, FL: Academic Press, Inc. 1993, pp. 98-181.
