EpiCypher recently launched CUTANA™ CUT&RUN assays, which generate high quality chromatin profiling data with reduced cell inputs and sequencing requirements compared with ChIP-seq. Due to the numerous advantages over ChIP-seq, CUT&RUN (and its sister technology, CUT&Tag) is quickly becoming the preferred approach for chromatin mapping experiments.
To supplement these advances, EpiCypher is also leading the validation of highly specific antibodies to histone PTMs for chromatin profiling assays. Notably, the ChIP field has been notoriously misled by improperly validated antibodies; even highly cited, “ChIP-grade” antibodies have been shown to display unacceptable performance (e.g. significant off-target binding, low efficiency, and/or significant lot-to-lot variation) 1-3. Read more about this topic here. To avoid these same pitfalls with CUT&RUN technology, the use of antibodies carefully validated for CUT&RUN performance is essential.
We previously developed SNAP-ChIP® Spike-in Technology to rigorously assess antibody specificity and efficiency for ChIP experiments using our modified recombinant nucleosomes (dNucs) as controls (see previous blog). We have further leveraged this technology to identify CUTANA-compatible antibodies for histone PTMs in CUT&RUN (such as H3K4me3), as we have found that many SNAP-ChIP Certified Antibodies also display robust performance in CUT&RUN assays.
While SNAP-ChIP excels at validating antibodies to histone PTMs, this approach is not yet compatible for non-PTM targets of interest. This leaves a major gap in the validation of high-value chromatin-interacting protein targets, such as transcription factors, epigenetic reader proteins, epigenetic enzymes, and chromatin remodeling proteins.
To identify best-in-class antibodies to chromatin associated proteins for CUT&RUN assays, EpiCypher has developed a rigorous pipeline to assess antibody specificity and performance. As described in more detail below, our validation pipeline includes:
- Testing multiple antibodies for the same target in CUT&RUN profiling assays. We then move forward with the top-performing antibodies that display both genome-wide and locus-specific enrichment recapitulating known target biology.
- Performing rigorous lot-specific QC.
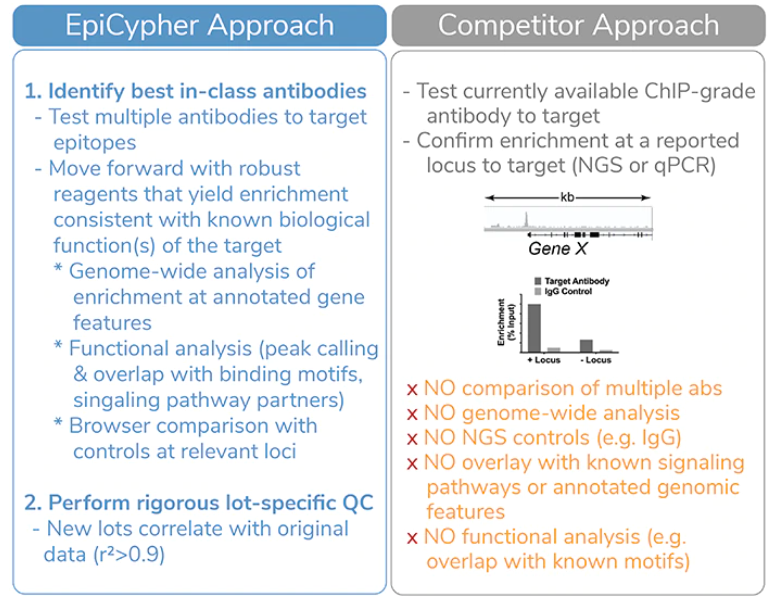
This approach is significantly more thorough than the standard “validation” approach utilized by competitor antibody providers (Figure 1). Our first set of CUTANA-compatible antibodies to non-histone PTM targets covers three important protein classes: transcription factors (FOXA1), chromatin readers (BRD4), and chromatin remodelers (BRG1/SMARCA4).
Figure 1: EpiCypher’s validation approach for CUT&RUN antibodies to chromatin-associated proteins is significantly more rigorous compared to the approach of our competitors.
1) Identification of top-performing antibodies with robust enrichment consistent with known biological function of target
The first step in our validation approach is to perform CUT&RUN using multiple antibodies for the target of interest. We compare the resulting enrichment patterns and eliminate antibodies with poor enrichment or that are clear outliers, moving forward only with antibodies that display high signal-to-noise and robust peaks that are consistent with the known biology of the target, both genome-wide and at specific loci of interest (e.g. analysis of DNA motifs, overlap with known signaling proteins).
Genome-wide enrichment at annotated features
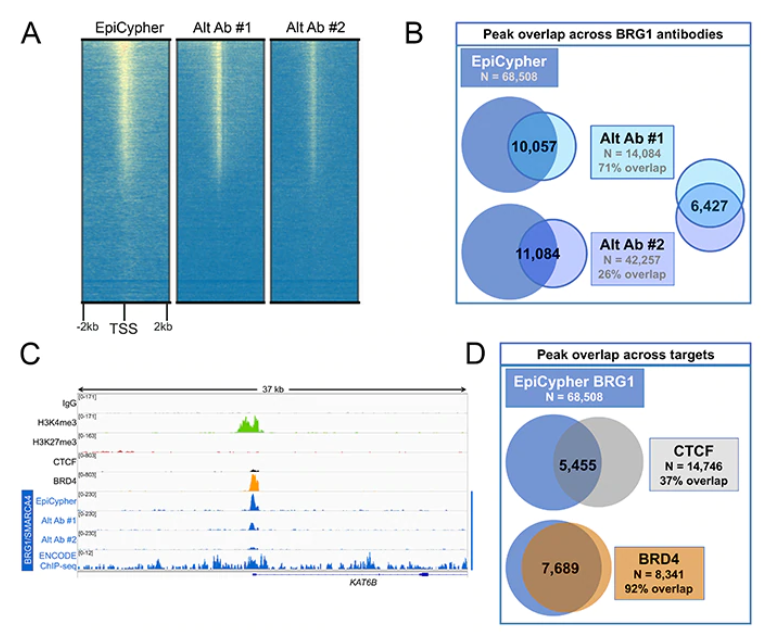
For example, to develop a best-in-class CUT&RUN-compatible antibody for the chromatin remodeler BRG1 (SMARCA4), we compared 3 different antibodies (Figure 2A). BRG1 is a component of the ATP-dependent chromatin remodeling complex SWI/SNF (BAF). SWI/SNF plays a crucial role in regulation of gene expression and differentiation, and mutations in this complex are implicated in various neurodevelopmental disorders and cancers 4. BRG1 has been shown to directly bind other important chromatin-interacting proteins, including the transcription factor CTCF 5 and the chromatin remodeler BRD4 6. Notably, BRG1 and BRD4 exhibit significant co-binding genome-wide; previous work reported over 80% overlap between BRD4 and BRG1 peaks 6. We leveraged our understanding of this biology to select the best-performing BRG1 antibody for CUT&RUN.
As displayed in Figures 2A and 2B, one BRG1 antibody (labeled EpiCypher) exhibited far superior performance, displaying the highest signal-to-noise and detecting the greatest number of peaks compared with the two alternate antibodies. While a portion of the peaks detected by this antibody were also detected by the alternate antibodies, as reflected by the regions of overlap, the reduced signal-to-noise of the alternate reagents resulted in far fewer peaks overall.
Figure 2: Comparison and validation of a BRG1 antibody. (A) CUT&RUN was performed using three BRG1 antibodies. Sequencing reads were aligned to annotated TSSs (+/- 2 kbp) of 18,793 genes. High, medium, and low signal is ranked by intensity (top to bottom), aligned relative to H3K4me3 antibody (not shown). (B) The EpiCypher antibody produced the largest number of peaks, a portion of which were also detected by the alternate antibodies. (C) EpiCypher antibody displays high signal-to-noise and detects a significant peak that is missed by alternate antibodies. (D) BRG1 peaks identified by EpiCypher antibody overlap with peaks for known interacting proteins. The lower and higher degrees of overlap for CTCF and BRD4, respectively, is consistent with literature.
Overlap with known binding partners
Importantly, as a next step following global enrichment analysis, we compared overlap of peaks between the top-performing BRG1 antibody and known BRG1-binding proteins CTCF and BRD4 5,6 (Figures 2C and 2D). The strong pattern of overlap with CTCF and BRD4 suggest that the EpiCypher CUTANA-compatible BRG1 antibody is highly specific. These data also support the conclusion that the increased signal compared with the two alternate antibodies is due to improved BRG1 detection, rather than off-target binding or background.
Motif analysis to examine on-target binding
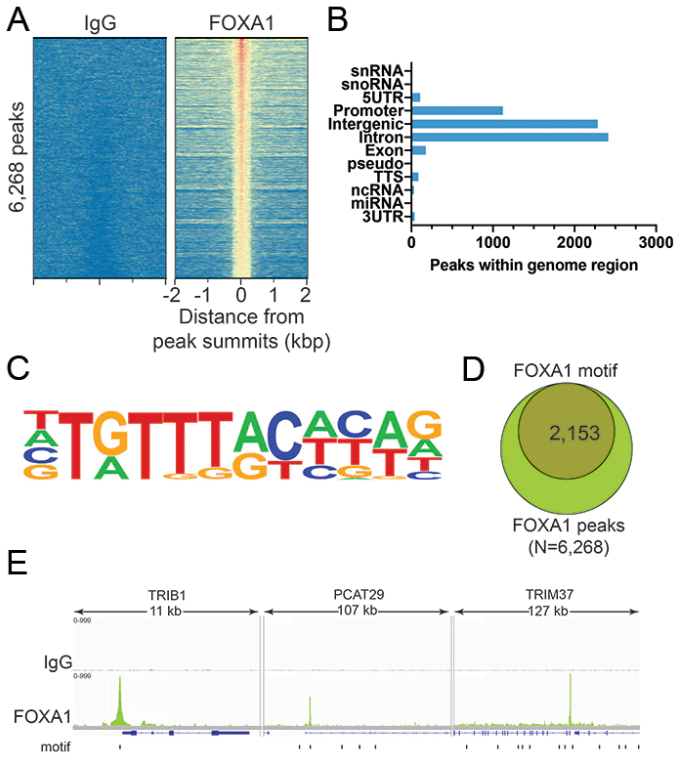
As noted above, many chromatin interacting proteins bind specific DNA sequences, which we also leveraged for CUTANA CUT&RUN antibody testing. FOXA1 belongs to a special set of transcription factors known as “pioneer factors,” which refers to their ability to directly bind and “open” condensed chromatin structure. Relaxation of heterochromatin by pioneer factors facilitates interactions with other chromatin-associated proteins, including transcription factors, chromatin remodeling complexes or modifying enzymes, to drive changes in gene expression. FOXA1 is particularly essential for embryonic development, including establishment of tissue-specific gene expression 7.
For our CUT&RUN-compatible antibody for the transcription factor FOXA1, we first verified high signal-to-noise in our CUT&RUN data and confirmed expected genome-wide distribution, as explained above for BRG1 (Figures 3A and 3B). We then proceeded to compare enrichment with known FOXA1 DNA-binding motifs and observed strong overlap with these motifs (Figures 3C-E). Together, these data confirm the reliability of our FOXA1 antibody for superior performance in CUT&RUN.
Figure 3: Validation of FOXA1 antibody. (A) Heatmap showing FOXA1 peaks relative to IgG negative control antibody in aligned rows ranked by intensity. (B) The number of peaks which fall into distinct classes of functionally annotated genomic regions is plotted. (C) HOMER software was used to identify known motifs underneath FOXA1 CUT&RUN peaks. The FOXA1 consensus motif was highly enriched, and is represented as a sequence logo position weight matrix. (D) Venn diagram representing the number of FOXA1 peaks containing FOXA1 consensus motifs. (E) Three representative loci showing FOXA1 peaks overlapping with FOXA1 consensus motifs (Integrative Genomics Viewer).
Lot-to-lot quality control
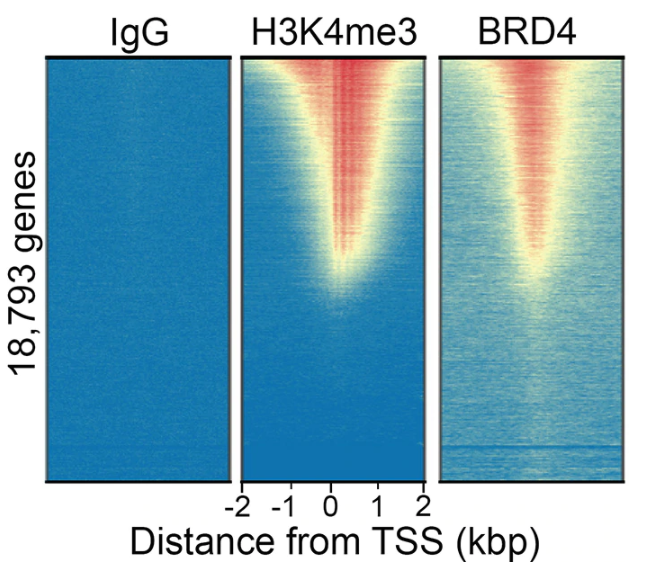
To ensure optimal and reliable performance of our antibodies over time, we independently validate each lot of antibody using the same analyses described above, and confirm consistent performance with original data (r >90%). We compare each lot of antibody at the genome-wide level to ensure localized enrichment with high signal-to-noise at the expected genomic features, as demonstrated by our validation of the BRD4 antibody shown in Figure 4. We also perform motif analysis and overlap of peaks generated by different lots of the same antibody. This lot-validation strategy is standard for all of our CUTANA CUT&RUN-compatible antibodies, including our antibodies targeting histone PTMs.
Figure 4: Genome-wide distribution of BRD4 is consistent with H3K4me3, with IgG negative control for comparison. Sequencing reads were aligned to annotated TSSs (+/- 2 kbp) of 18,793 genes. High, medium, and low signal is ranked by intensity (top to bottom), aligned relative to H3K4me3.
Conduct your CUT&RUN experiments with confidence
Antibody choice shouldn’t be left to chance. EpiCypher’s CUTANA CUT&RUN compatible antibodies are best-in-class reagents, having been extensively evaluated on a lot-by-lot basis to ensure they are specific and reliable detection reagents for CUT&RUN assays.
Interested in using CUTANA-compatible antibodies in your CUT&RUN experiments? More targets and target classes (e.g. epigenetic enzymes), as well as antibodies validated for CUT&Tag assays, are coming soon! Follow EpiCypher on Twitter and LinkedIn for updates.