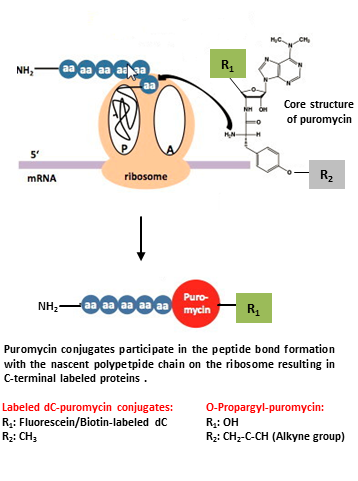
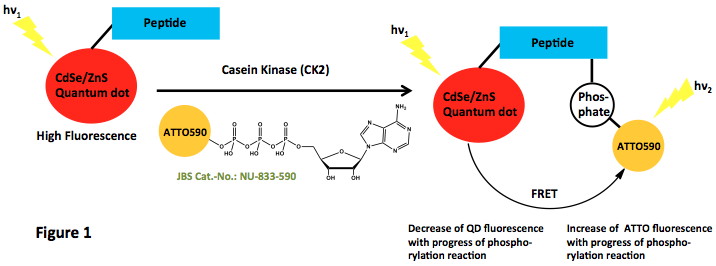
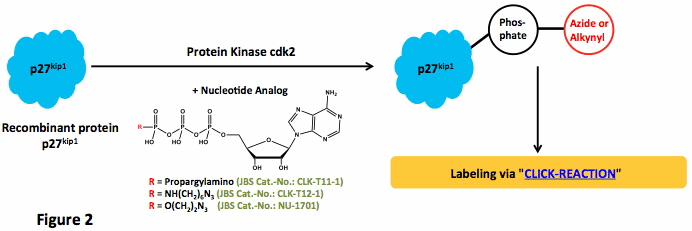
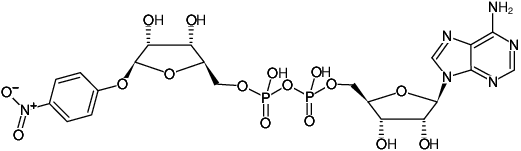
Probes for Protein Kinases and Protein Phosphatases
Protein phosphorylation is a regulatory mechanism that is extremely important in most cellular processes such as protein synthesis, cell division, signal transduction, cell growth, development and aging as many enzymes and receptors are activated and deactivated via phosphorylation/dephosphorylation due to specific protein kinases and protein phosphatases[1-4].
Phosphorylation and dephosphorylation can modify the function of a protein in almost every conceivable way; for example by increasing or decreasing its biological activity, by stabilizing it or marking it for destruction, by facilitating or inhibiting movement between subcellular compartments, or by initiating or disrupting protein– protein interactions. It is believed that about 30% of the proteins encoded by the human genome contain covalently bound phosphate and abnormal phosphorylation is recognized as a cause or consequence of many human diseases as well[5].
Jena Bioscience offers a selection of modified biomolecules and small-molecule reporters such as labeled ATPs as phosphoryl donor to analyze the complex processes of protein phosphorylation and dephosphorylation.
| Name | Cat. No. | Size |
| DDAO Phosphate | APC-001-1 | 1 mg |
| DDAO Phosphate | APC-001-5 | 5 x 1 mg |
| γ-[(PEG3-Amino)-imido]-ATP-Biotin | NU-970-BIO | 50 μl (1 mM) |
| γ-(6-Aminohexyl)-ATP-ATTO-590 | NU-833-590 | 80 μl (1 mM) |
| γ-[(Propargyl)-imido]-ATP | CLK-T11-1 | 1 mg |
| γ-[(6-Azidohexyl)-imido]-ATP | CLK-T12-1 | 1 mg |
| γ-(2-Azidoethyl)-ATP | NU-1701S | 100 μl (10 mM) |
| γ-(2-Azidoethyl)-ATP | NU-1701L | 5 x 100 μl (10 mM) |
| ATPγS | NU-406-5 | 5 mg |
| ATPγS | NU-406-25 | 25 mg |
| ATPγS | NU-406-50 | 50 mg |
| ATP-acetyl-hex-Biotin | NU-277 | 16 x 0.01 μmol (16 x approximately 11.5 μg) |
| ATP-acetyl-Desthiobiotin | NU-276 | 16 x 0.01 μmol (16 x approximately 10.1 μg) |