Chemical Release
Hydrazinolysis is suitable for releasing both N- and O-linked glycans from glycoproteins. It involves the incubation of the dried glycoprotein with anhydrous hydrazine followed by a straightforward workup to purify the released glycans. For success, hydrazinolysis must be carried out with absolutely pure hydrazine under strictly controlled conditions. Advantages of hydrazinolysis include its versatility with the ability to release both N- and O-linked glycans in a virtually non-selective way.
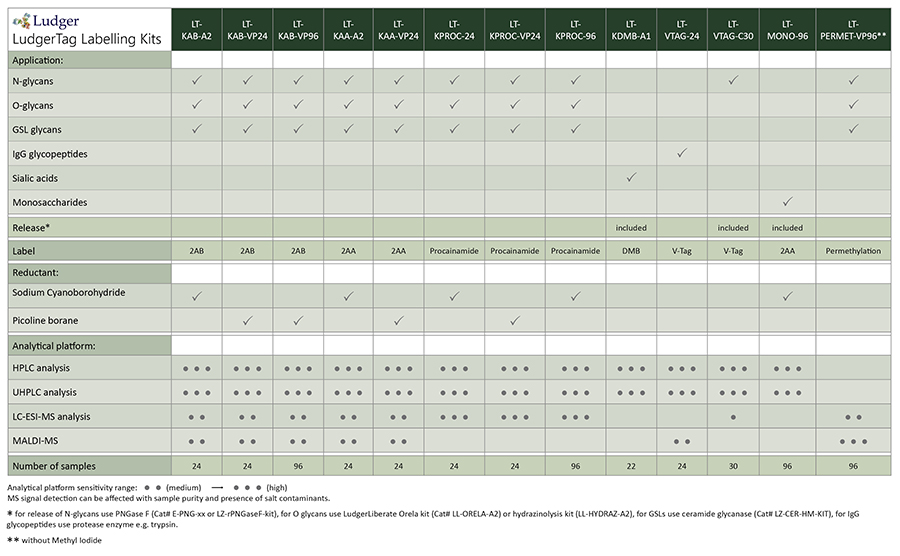
Ludger products for chemical release of glycans:
Hydrazinolysis kit (LL-HYDRAZ-A2): For release of N + O-linked glycans. Released glycans have free reducing terminii to allow fluorescent tagging by reductive amination. The release conditions can be optimized for release of N-glycans, O-glycans or both N- and O-glycans.
Orela kit (LL-ORELA-A2): For release of O-linked glycans. Released glycans have free reducing terminii to allow fluorescent tagging by reductive amination.
Enzyme (endoglycosidase) Release
Endoglycosidase release is generally a simpler method to implement than hydrazinolysis and suitable for many applications. There are a number of useful endoglycosidases, including Peptide N Glycosidase F (PNGase F), which releases most N-glycans from glycoproteins. Take care when using endoglycosidases as there are a number of conditions and substances that can lead to selective non-release of glycans. In particular, PNGaseF does not release N-glycans which have core fucose attached in the alpha1-3 position which can be found on N-glycans from plants and insects.
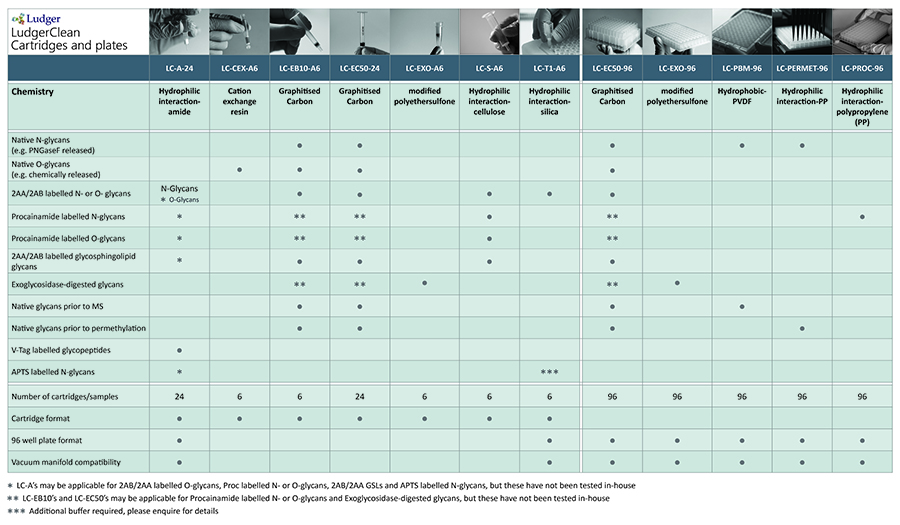
Ludger products for chemical release of glycans:
PNGase F (E-PNG-01 or E-PNG01-200): removes N glycans.
O-Glycosidase (E-G001): removes core 1 O glycans (Gal-β(1-3)GalNAc- alpha attached to the serine or threonine)
LudgerZyme Ceramide Glycanase kit (LZ-CER-HM-KIT): Ceramide glycanase deglycosylates a variety of glycosphingolipids by cleaving the β-glycosyl linkage.