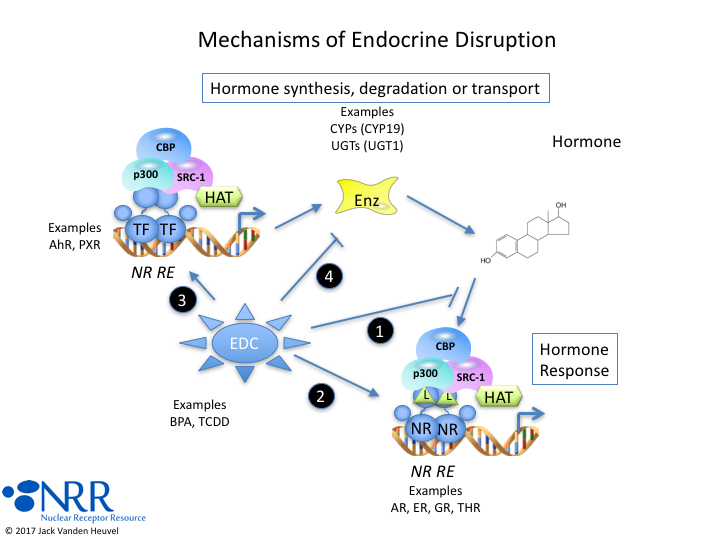
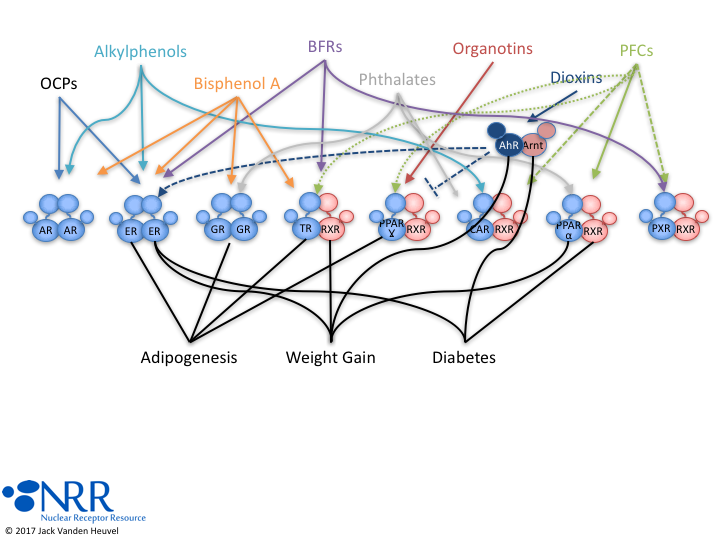
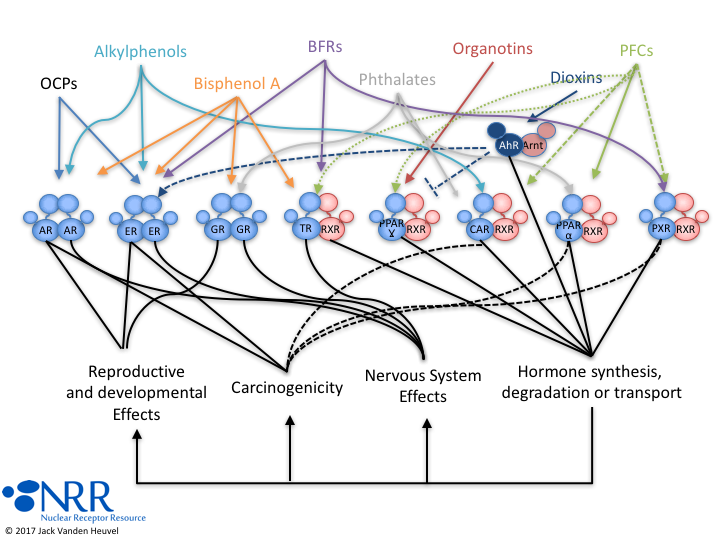
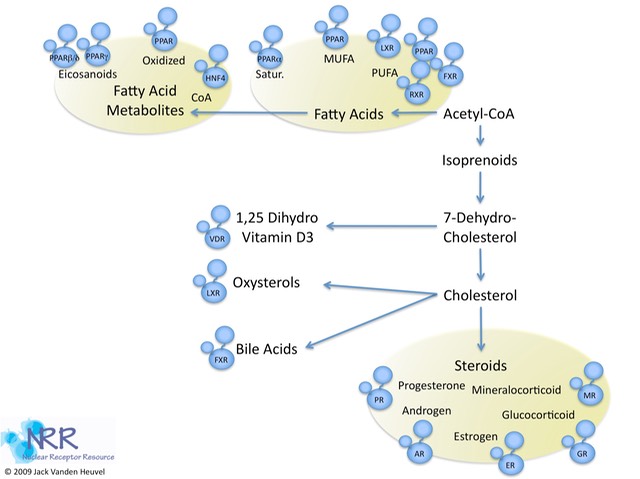
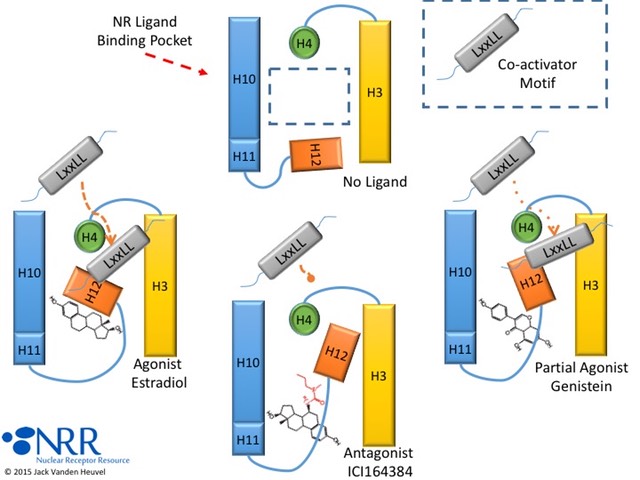
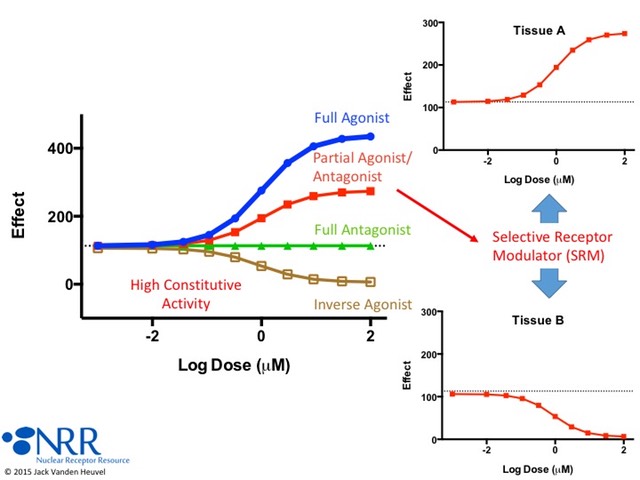
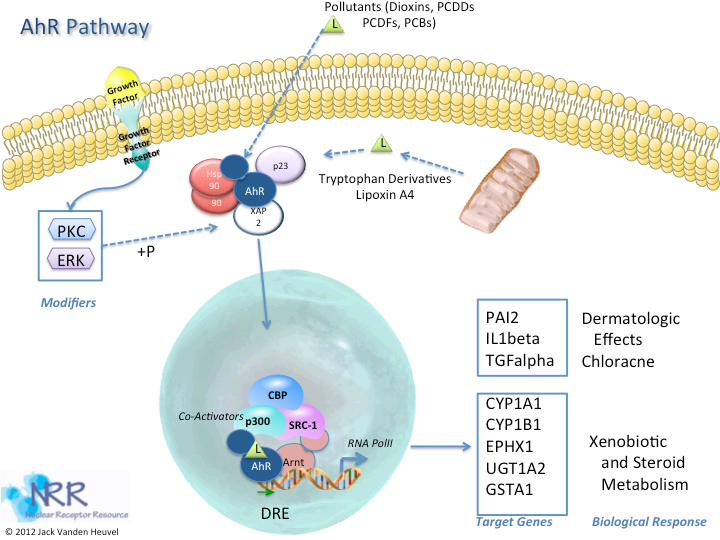
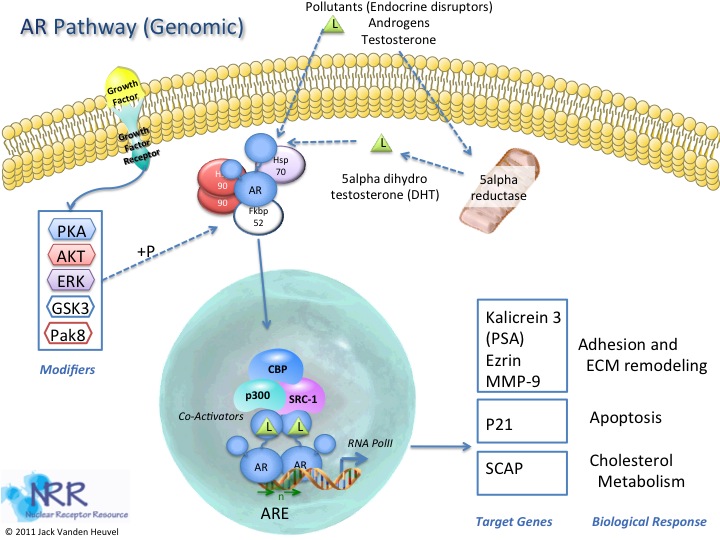
| Description |
Sizes |
Catalog # |
Species |
Target Type |
Associated Disease |
| CB1R Reporter Assay Kit |
1 x 96-well format assays |
IB19001 |
Human |
GPCR |
Cancer, Cardiovascular, Dermatitis, Dyslipidemia, NASH/NAFLD, Neurodegenerative, Obesity, Osteoporosis |
| CB1R Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB19001-32 |
Human |
GPCR |
Cancer, Cardiovascular, Dermatitis, Dyslipidemia, NASH/NAFLD, Neurodegenerative, Obesity, Osteoporosis |
| CB1R Reporter Assay Kit |
1 x 384-well format assays |
IB19002 |
Human |
GPCR |
Cancer, Cardiovascular, Dermatitis, Dyslipidemia, NASH/NAFLD, Neurodegenerative, Obesity, Osteoporosis |
| Cyn Monkey FXR Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
C00601 |
Cyn Monkey |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Reproduction |
| Cyn Monkey PGR Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
C05001 |
Cyn Monkey |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Obesity, Osteoporosis, Reproduction |
| Cyn Monkey PPARa Reporter Assay Kit |
1 x 96-well format assayst / 3 x 32 assays in 96-well format |
C00111 |
Cyn Monkey |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cancer, Cardiovascular, Dermatitis, Dyslipidemia, NASH/NAFLD, Obesity, Reproduction |
| Cyn Monkey PPARd Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
C00121 |
Cyn Monkey |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cancer, Dermatitis, Dyslipidemia, NASH/NAFLD, Neurodegenerative, Obesity, Reproduction, Wound Healing |
| Cyn Monkey PPARg Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
C00101 |
Cyn Monkey |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Osteoporosis, Reproduction |
| Cyn Monkey PXR Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
C07001 |
Cyn Monkey |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cancer, Cardiovascular, Kidney Disease, NASH/NAFLD, Wound Healing |
| Dog FXR Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
D00601 |
Dog |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Reproduction |
| Dog LXRa Reporter Assay Kit |
1 x 96-well format assays |
D00311 |
Dog |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Autoimmune, Cancer, Cardiovascular, NASH/NAFLD, Reproduction |
| Dog LXRb Reporter Assay Kit |
1 x 96-well format assays |
D00301 |
Dog |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cancer, Cardiovascular, Dyslipidemia, Reproduction |
| Dog PPARa Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
D00111 |
Dog |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cancer, Cardiovascular, Dermatitis, Dyslipidemia, NASH/NAFLD, Obesity, Reproduction |
| Dog PPARd Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
D00121 |
Dog |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cancer, Dermatitis, Dyslipidemia, NASH/NAFLD, Neurodegenerative, Obesity, Reproduction, Wound Healing |
| Dog PXR Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
D07001 |
Dog |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cancer, Cardiovascular, Kidney Disease, NASH/NAFLD, Wound Healing |
| EPOR Reporter Assay Kit |
1 x 96-well format assays |
IB17001 |
Human |
Cytokine Receptor |
Cancer, Kidney Disease, Neurodegenerative, Reproduction |
| EPOR Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB17001-32 |
Human |
Cytokine Receptor |
Cancer, Kidney Disease, Neurodegenerative, Reproduction |
| EPOR Reporter Assay Kit |
1 x 384-well format assays |
IB17002 |
Human |
Cytokine Receptor |
Cancer, Kidney Disease, Neurodegenerative, Reproduction |
| Expression Profiling of Clinically Relevant CYPs Assay Kit |
2 x 48 assays in 96-well format |
UGE1003-48 |
Human |
|
|
| FGFR/β-Klotho Reporter Assay Kit |
1 x 96-well format assays |
IB22001 |
Human |
Growth Factor Receptor |
Cardiovascular, NASH/NAFLD, Obesity, Reproduction, Wound Healing |
| FGFR/β-Klotho Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB22001-32 |
Human |
Growth Factor Receptor |
Cardiovascular, NASH/NAFLD, Obesity, Reproduction, Wound Healing |
| FGFR/β-Klotho Reporter Assay Kit |
1 x 384-well format assays |
IB22002 |
Human |
Growth Factor Receptor |
Cardiovascular, NASH/NAFLD, Obesity, Reproduction, Wound Healing |
| FGFR1/2 Reporter Assay Kit |
1 x 96-well format assays |
IB21001 |
Human |
Growth Factor Receptor |
Autoimmune, Cancer, Kidney Disease, NASH/NAFLD, Neurodegenerative, Reproduction, Wound Healing |
| FGFR1/2 Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB21001-32 |
Human |
Growth Factor Receptor |
Autoimmune, Cancer, Kidney Disease, NASH/NAFLD, Neurodegenerative, Reproduction, Wound Healing |
| FGFR1/2 Reporter Assay Kit |
1 x 384-well format assays |
IB21002 |
Human |
Growth Factor Receptor |
Autoimmune, Cancer, Kidney Disease, NASH/NAFLD, Neurodegenerative, Reproduction, Wound Healing |
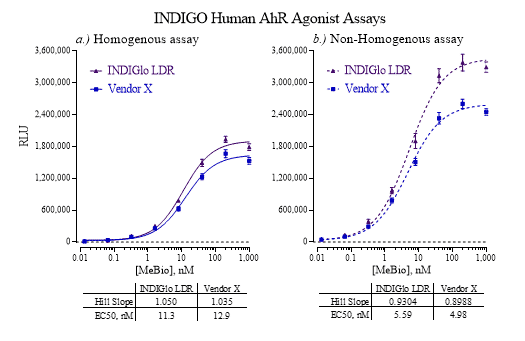
| Human AhR Reporter Assay Kit |
1 x 96-well format assays |
IB06001 |
Human |
Nuclear Hormone Receptor |
Cancer, Cardiovascular, Dermatitis, Kidney Disease, NASH/NAFLD, Reproduction, Wound Healing |
| Human AhR Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB06001-32 |
Human |
Nuclear Hormone Receptor |
Cancer, Cardiovascular, Dermatitis, Kidney Disease, NASH/NAFLD, Reproduction, Wound Healing |
| Human AhR Reporter Assay Kit |
1 x 384-well format assays |
IB06002 |
Human |
Nuclear Hormone Receptor |
Cancer, Cardiovascular, Dermatitis, Kidney Disease, NASH/NAFLD, Reproduction, Wound Healing |
| Human AR Reporter Assay Kit |
1 x 96-well format assays |
IB03001 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, Dyslipidemia, Kidney Disease, NASH/NAFLD, Neurodegenerative, Obesity, Osteoporosis, Reproduction |
| Human AR Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB03001-32 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, Dyslipidemia, Kidney Disease, NASH/NAFLD, Neurodegenerative, Obesity, Osteoporosis, Reproduction |
| Human AR Reporter Assay Kit |
1 x 384-well format assays |
IB03002 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, Dyslipidemia, Kidney Disease, NASH/NAFLD, Neurodegenerative, Obesity, Osteoporosis, Reproduction |
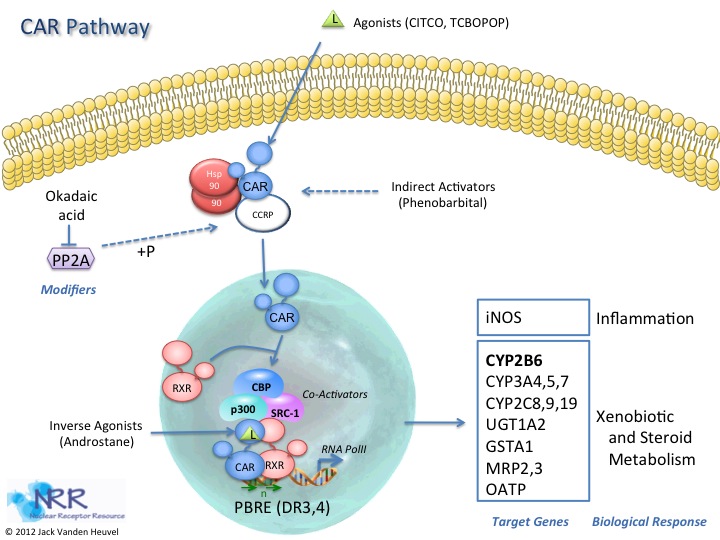
| Human CAR-2 Reporter Assay Kit |
1 x 96-well format assays |
IB00921 |
Human |
Nuclear Hormone Receptor |
NASH/NAFLD, Wound Healing |
| Human CAR-2 Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB00921-32 |
Human |
Nuclear Hormone Receptor |
NASH/NAFLD, Wound Healing |
| Human CAR-2 Reporter Assay Kit |
1 x 384-well format assays |
IB00922 |
Human |
Nuclear Hormone Receptor |
NASH/NAFLD, Wound Healing |
| Human CAR-3 Reporter Assay Kit |
1 x 96-well format assays |
IB00901 |
Human |
Nuclear Hormone Receptor |
NASH/NAFLD, Wound Healing |
| Human CAR-3 Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB00901-32 |
Human |
Nuclear Hormone Receptor |
NASH/NAFLD, Wound Healing |
| Human CAR-3 Reporter Assay Kit |
1 x 384-well format assays |
IB00902 |
Human |
Nuclear Hormone Receptor |
NASH/NAFLD, Wound Healing |
| Human EGFR1 Reporter Assay Kit |
1 x 96-well format assays |
IB13001 |
Human |
Growth Factor Receptor |
Cancer, Dermatitis, Kidney Disease, NASH/NAFLD, Neurodegenerative, Obesity, Reproduction, Wound Healing |
| Human EGFR1 Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB13001-32 |
Human |
Growth Factor Receptor |
Cancer, Dermatitis, Kidney Disease, NASH/NAFLD, Neurodegenerative, Obesity, Reproduction, Wound Healing |
| Human EGFR1 Reporter Assay Kit |
1 x 384-well format assays |
IB13002 |
Human |
Growth Factor Receptor |
Cancer, Dermatitis, Kidney Disease, NASH/NAFLD, Neurodegenerative, Obesity, Reproduction, Wound Healing |
| Human ERa Reporter Assay Kit |
1 x 96-well format assays |
IB00401 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cardiovascular, Dyslipidemia, NASH/NAFLD, Obesity, Osteoporosis, Reproduction, Wound Healing |
| Human ERa Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB00401-32 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cardiovascular, Dyslipidemia, NASH/NAFLD, Obesity, Osteoporosis, Reproduction, Wound Healing |
| Human ERa Reporter Assay Kit |
1 x 384-well format assays |
IB00402 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cardiovascular, Dyslipidemia, NASH/NAFLD, Obesity, Osteoporosis, Reproduction, Wound Healing |
| Human ERb Reporter Assay Kit |
1 x 96-well format assays |
IB00411 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, Neurodegenerative, Obesity, Osteoporosis, Reproduction, Wound Healing |
| Human ERb Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB00411-32 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, Neurodegenerative, Obesity, Osteoporosis, Reproduction, Wound Healing |
| Human ERb Reporter Assay Kit |
1 x 384-well format assays |
IB00412 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, Neurodegenerative, Obesity, Osteoporosis, Reproduction, Wound Healing |
| Human ERRa Reporter Assay Kit |
1 x 96-well format assays |
IB08001 |
Human |
Nuclear Hormone Receptor |
Cancer |
| Human ERRa Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB08001-32 |
Human |
Nuclear Hormone Receptor |
Cancer |
| Human ERRa Reporter Assay Kit |
1 x 384-well format assays |
IB08002 |
Human |
Nuclear Hormone Receptor |
Cancer |
| Human ERRb Reporter Assay Kit |
1 x 96-well format assays |
IB08011 |
Human |
Nuclear Hormone Receptor |
Cancer |
| Human ERRb Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB08011-32 |
Human |
Nuclear Hormone Receptor |
Cancer |
| Human ERRb Reporter Assay Kit |
1 x 384-well format assays |
IB08012 |
Human |
Nuclear Hormone Receptor |
Cancer |
| Human ERRg Reporter Assay Kit |
1 x 96-well format assays |
IB08021 |
Human |
Nuclear Hormone Receptor |
Cancer |
| Human ERRg Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB08021-32 |
Human |
Nuclear Hormone Receptor |
Cancer |
| Human ERRg Reporter Assay Kit |
1 x 384-well format assays |
IB08022 |
Human |
Nuclear Hormone Receptor |
Cancer |
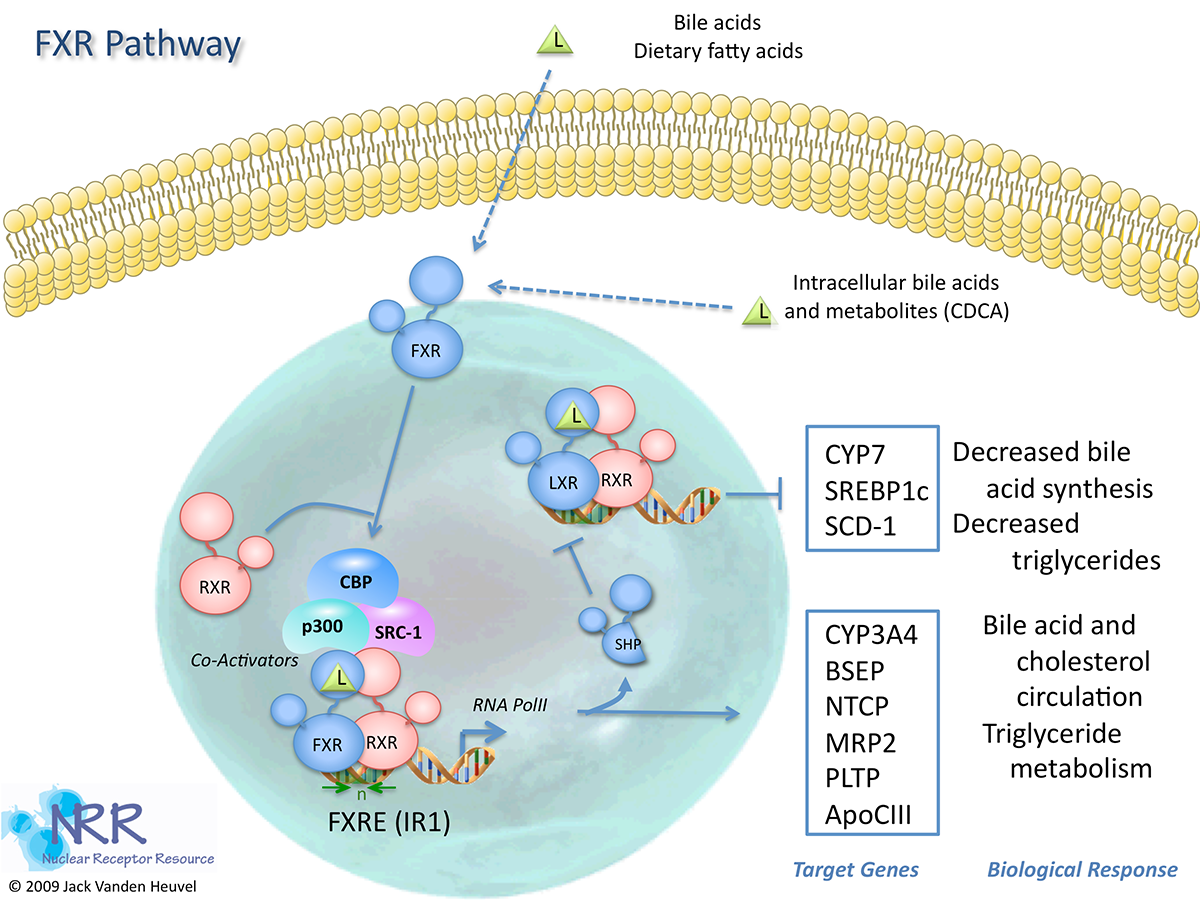
| Human FXR Reporter Assay Kit |
1 x 96-well format assays |
IB00601 |
Human |
Nuclear Hormone Receptor |
Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Reproduction |
| Human FXR Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB00601-32 |
Human |
Nuclear Hormone Receptor |
Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Reproduction |
| Human FXR Reporter Assay Kit |
1 x 384-well format assays |
IB00602 |
Human |
Nuclear Hormone Receptor |
Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Reproduction |
| Human GHR Reporter Assay Kit |
1 x 96-well format assays |
IB14001 |
Human |
Growth Factor Receptor |
Autoimmune, Cancer, Cardiovascular, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Osteoporosis, Reproduction |
| Human GHR Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB14001-32 |
Human |
Growth Factor Receptor |
Autoimmune, Cancer, Cardiovascular, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Osteoporosis, Reproduction |
| Human GHR Reporter Assay Kit |
1 x 384-well format assays |
IB14002 |
Human |
Growth Factor Receptor |
Autoimmune, Cancer, Cardiovascular, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Osteoporosis, Reproduction |
| Human GR Reporter Assay Kit |
1 x 96-well format assays |
IB00201 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Osteoporosis, Reproduction |
| Human GR Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB00201-32 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Osteoporosis, Reproduction |
| Human GR Reporter Assay Kit |
1 x 384-well format assays |
IB00202 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Osteoporosis, Reproduction |
| Human IGF-1R Reporter Assay Kit |
1 x 96-well format assays |
IB29001 |
Human |
Growth Factor Receptor |
Cancer, Obesity, Reproduction |
| Human IGF-1R Reporter Assay Kit |
1 x 384-well format assays |
IB29002 |
Human |
Growth Factor Receptor |
Cancer, Obesity, Reproduction |
| Human INSRb Reporter Assay Kit |
1 x 96-well format assays |
IB28001 |
Human |
Growth Factor Receptor |
Obesity, Reproduction |
| Human INSRb Reporter Assay Kit |
1 x 384-well format assays |
IB28002 |
Human |
Growth Factor Receptor |
Obesity, Reproduction |
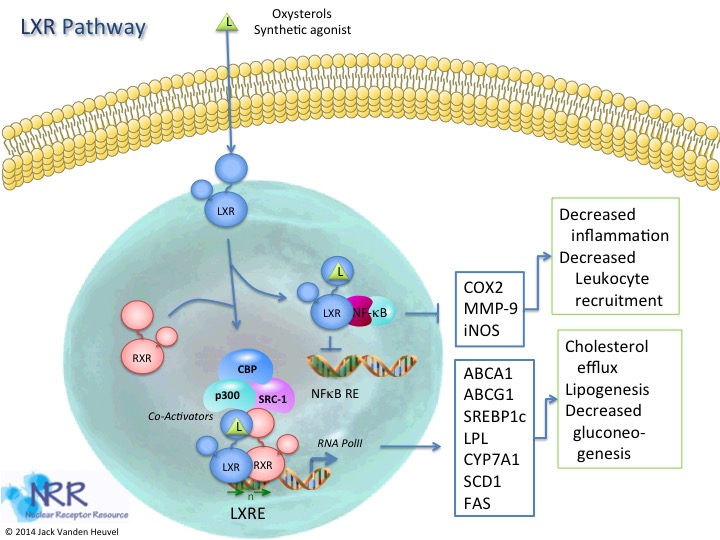
| Human LXRa Reporter Assay Kit |
1 x 96-well format assays |
IB00311 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, NASH/NAFLD, Reproduction |
| Human LXRa Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB00311-32 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, NASH/NAFLD, Reproduction |
| Human LXRa Reporter Assay Kit |
1 x 384-well format assays |
IB00312 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, NASH/NAFLD, Reproduction |
| Human LXRb Reporter Assay Kit |
1 x 96-well format assays |
IB00301 |
Human |
Nuclear Hormone Receptor |
Cancer, Cardiovascular, Dyslipidemia, Reproduction |
| Human LXRb Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB00301-32 |
Human |
Nuclear Hormone Receptor |
Cancer, Cardiovascular, Dyslipidemia, Reproduction |
| Human LXRb Reporter Assay Kit |
1 x 384-well format assays |
IB00302 |
Human |
Nuclear Hormone Receptor |
Cancer, Cardiovascular, Dyslipidemia, Reproduction |
| Human MR Reporter Assay Kit |
1 x 96-well format assays |
IB00501 |
Human |
Nuclear Hormone Receptor |
Cardiovascular, Dyslipidemia, Kidney Disease, Osteoporosis |
| Human MR Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB00501-32 |
Human |
Nuclear Hormone Receptor |
Cardiovascular, Dyslipidemia, Kidney Disease, Osteoporosis |
| Human MR Reporter Assay Kit |
1 x 384-well format assays |
IB00502 |
Human |
Nuclear Hormone Receptor |
Cardiovascular, Dyslipidemia, Kidney Disease, Osteoporosis |
| Human P-Glycoprotein / MDR1 Drug Interaction Assay |
2 x 48 format |
HPGP-48 |
Human |
|
|
| Human PDGFR a/b Reporter Assay Kit |
1 x 96-well format assays |
IB23001 |
Human |
Growth Factor Receptor |
Wound Healing |
| Human PDGFR a/b Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB23001-32 |
Human |
Growth Factor Receptor |
Wound Healing |
| Human PDGFR a/b Reporter Assay Kit |
1 x 384-well format assays |
IB23002 |
Human |
Growth Factor Receptor |
Wound Healing |
| Human PGR Reporter Assay Kit |
1 x 96-well format assays |
IB05001 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Obesity, Osteoporosis, Reproduction |
| Human PGR Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB05001-32 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Obesity, Osteoporosis, Reproduction |
| Human PGR Reporter Assay Kit |
1 x 384-well format assays |
IB05002 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Obesity, Osteoporosis, Reproduction |
| Human PPARa Reporter Assay Kit |
1 x 96-well format assays |
IB00111 |
Human |
Nuclear Hormone Receptor |
Cancer, Cardiovascular, Dermatitis, Dyslipidemia, NASH/NAFLD, Obesity, Reproduction |
| Human PPARa Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB00111-32 |
Human |
Nuclear Hormone Receptor |
Cancer, Cardiovascular, Dermatitis, Dyslipidemia, NASH/NAFLD, Obesity, Reproduction |
| Human PPARa Reporter Assay Kit |
1 x 384-well format assays |
IB00112 |
Human |
Nuclear Hormone Receptor |
Cancer, Cardiovascular, Dermatitis, Dyslipidemia, NASH/NAFLD, Obesity, Reproduction |
| Human PPARd Reporter Assay Kit |
1 x 96-well format assays |
IB00121 |
Human |
Nuclear Hormone Receptor |
Cancer, Dermatitis, Dyslipidemia, NASH/NAFLD, Neurodegenerative, Obesity, Reproduction, Wound Healing |
| Human PPARd Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB00121-32 |
Human |
Nuclear Hormone Receptor |
Cancer, Dermatitis, Dyslipidemia, NASH/NAFLD, Neurodegenerative, Obesity, Reproduction, Wound Healing |
| Human PPARd Reporter Assay Kit |
1 x 384-well format assays |
IB00122 |
Human |
Nuclear Hormone Receptor |
Cancer, Dermatitis, Dyslipidemia, NASH/NAFLD, Neurodegenerative, Obesity, Reproduction, Wound Healing |
| Human PPARg Reporter Assay Kit |
1 x 96-well format assays |
IB00101 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Osteoporosis, Reproduction |
| Human PPARg Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB00101-32 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Osteoporosis, Reproduction |
| Human PPARg Reporter Assay Kit |
1 x 384-well format assays |
IB00102 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Osteoporosis, Reproduction |
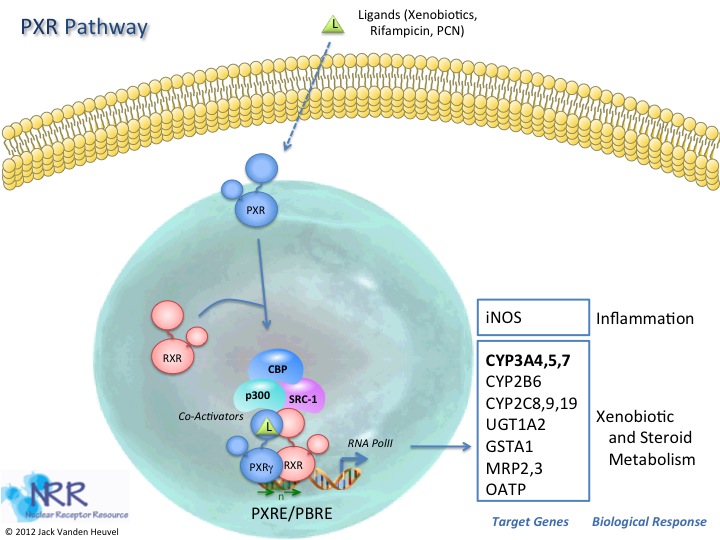
| Human PXR Reporter Assay Kit |
1 x 96-well format assays |
IB07001 |
Human |
Nuclear Hormone Receptor |
Cancer, Cardiovascular, Kidney Disease, NASH/NAFLD, Wound Healing |
| Human PXR Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB07001-32 |
Human |
Nuclear Hormone Receptor |
Cancer, Cardiovascular, Kidney Disease, NASH/NAFLD, Wound Healing |
| Human PXR Reporter Assay Kit |
1 x 384-well format assays |
IB07002 |
Human |
Nuclear Hormone Receptor |
Cancer, Cardiovascular, Kidney Disease, NASH/NAFLD, Wound Healing |
| Human RARa Reporter Assay Kit |
1 x 96-well format assays |
IB02201 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, Reproduction, Wound Healing |
| Human RARa Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB02201-32 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, Reproduction, Wound Healing |
| Human RARa Reporter Assay Kit |
1 x 384-well format assays |
IB02202 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, Reproduction, Wound Healing |
| Human RARb Reporter Assay Kit |
1 x 96-well format assays |
IB02101 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, Reproduction, Wound Healing |
| Human RARb Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB02101-32 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, Reproduction, Wound Healing |
| Human RARb Reporter Assay Kit |
1 x 384-well format assays |
IB02102 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, Reproduction, Wound Healing |
| Human RARg Reporter Assay Kit |
1 x 96-well format assays |
IB02001 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, Reproduction, Wound Healing |
| Human RARg Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB02001-32 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, Reproduction, Wound Healing |
| Human RARg Reporter Assay Kit |
1 x 384-well format assays |
IB02002 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, Reproduction, Wound Healing |
| Human RORa Reporter Assay Kit |
1 x 96-well format assays |
IB04011 |
Human |
Nuclear Hormone Receptor |
Cancer, Dyslipidemia, NASH/NAFLD |
| Human RORa Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB04011-32 |
Human |
Nuclear Hormone Receptor |
Cancer, Dyslipidemia, NASH/NAFLD |
| Human RORa Reporter Assay Kit |
1 x 384-well format assays |
IB04012 |
Human |
Nuclear Hormone Receptor |
Cancer, Dyslipidemia, NASH/NAFLD |
| Human RORg Reporter Assay Kit |
1 x 96-well format assays |
IB04001 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, NASH/NAFLD, Obesity |
| Human RORg Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB04001-32 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, NASH/NAFLD, Obesity |
| Human RORg Reporter Assay Kit |
1 x 384-well format assays |
IB04002 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, NASH/NAFLD, Obesity |
| Human RXRa Reporter Assay Kit |
1 x 96-well format assays |
IB00801 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, Reproduction, Wound Healing |
| Human RXRa Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB00801-32 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, Reproduction, Wound Healing |
| Human RXRa Reporter Assay Kit |
1 x 384-well format assays |
IB00802 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, Reproduction, Wound Healing |
| Human RXRb Reporter Assay Kit |
1 x 96-well format assays |
IB00811 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, Reproduction, Wound Healing |
| Human RXRb Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB00811-32 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, Reproduction, Wound Healing |
| Human RXRb Reporter Assay Kit |
1 x 384-well format assays |
IB00812 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, Reproduction, Wound Healing |
| Human RXRg Reporter Assay Kit |
1 x 96-well format assays |
IB00821 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, Dyslipidemia, Obesity, Wound Healing |
| Human RXRg Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB00821-32 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, Dyslipidemia, Obesity, Wound Healing |
| Human RXRg Reporter Assay Kit |
1 x 384-well format assays |
IB00822 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, Dyslipidemia, Obesity, Wound Healing |
| Human TEAD4/YAP Assay Kit |
1 x 96-well format assays |
IB16001 |
Human |
Transcription Factor |
Cancer, Cardiovascular, Kidney Disease, Neurodegenerative, Obesity, Reproduction |
| Human TEAD4/YAP Assay Kit |
3 x 32 assays in 96-well format |
IB16001-32 |
Human |
Transcription Factor |
Cancer, Cardiovascular, Kidney Disease, Neurodegenerative, Obesity, Reproduction |
| Human TEAD4/YAP Assay Kit |
1 x 384-well format assays |
IB16002 |
Human |
Transcription Factor |
Cancer, Cardiovascular, Kidney Disease, Neurodegenerative, Obesity, Reproduction |
| Human TGFbR Reporter Assay Kit |
1 x 96-well format assays |
IB12001 |
Human |
Growth Factor Receptor |
Autoimmune, Cancer, Cardiovascular, Neurodegenerative, Obesity, Wound Healing |
| Human TGFbR Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB12001-32 |
Human |
Growth Factor Receptor |
Autoimmune, Cancer, Cardiovascular, Neurodegenerative, Obesity, Wound Healing |
| Human TGFbR Reporter Assay Kit |
1 x 384-well format assays |
IB12002 |
Human |
Growth Factor Receptor |
Autoimmune, Cancer, Cardiovascular, Neurodegenerative, Obesity, Wound Healing |
| Human TGR5 Reporter Assay Kit |
1 x 96-well format assays |
IB26001 |
Human |
GPCR |
Cardiovascular |
| Human TGR5 Reporter Assay Kit |
1 x 384-well format assays |
IB26002 |
Human |
GPCR |
Cardiovascular |
| Human TRa Reporter Assay Kit |
1 x 96-well format assays |
IB01001 |
Human |
Nuclear Hormone Receptor |
Cancer, Cardiovascular, Dyslipidemia, NASH/NAFLD, Neurodegenerative, Obesity, Osteoporosis, Reproduction |
| Human TRa Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB01001-32 |
Human |
Nuclear Hormone Receptor |
Cancer, Cardiovascular, Dyslipidemia, NASH/NAFLD, Neurodegenerative, Obesity, Osteoporosis, Reproduction |
| Human TRa Reporter Assay Kit |
1 x 384-well format assays |
IB01002 |
Human |
Nuclear Hormone Receptor |
Cancer, Cardiovascular, Dyslipidemia, NASH/NAFLD, Neurodegenerative, Obesity, Osteoporosis, Reproduction |
| Human TRb Reporter Assay Kit |
1 x 96-well format assays |
IB01101 |
Human |
Nuclear Hormone Receptor |
Cancer, Cardiovascular, Dyslipidemia, NASH/NAFLD, Neurodegenerative, Obesity, Osteoporosis, Reproduction |
| Human TRb Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB01101-32 |
Human |
Nuclear Hormone Receptor |
Cancer, Cardiovascular, Dyslipidemia, NASH/NAFLD, Neurodegenerative, Obesity, Osteoporosis, Reproduction |
| Human TRb Reporter Assay Kit |
1 x 384-well format assays |
IB01102 |
Human |
Nuclear Hormone Receptor |
Cancer, Cardiovascular, Dyslipidemia, NASH/NAFLD, Neurodegenerative, Obesity, Osteoporosis, Reproduction |
| Human TrkA Reporter Assay Kit |
1 x 96-well format assays |
IB27011 |
Human |
Growth Factor Receptor |
Autoimmune, Cancer, Obesity |
| Human TrkA Reporter Assay Kit |
1 x 384-well format assays |
IB27012 |
Human |
Growth Factor Receptor |
Autoimmune, Cancer, Obesity |
| Human TrkB Reporter Assay Kit |
1 x 96-well format assays |
IB27021 |
Human |
Growth Factor Receptor |
Autoimmune, Cancer, Neurodegenerative, Reproduction |
| Human TrkB Reporter Assay Kit |
1 x 384-well format assays |
IB27022 |
Human |
Growth Factor Receptor |
Autoimmune, Cancer, Neurodegenerative, Reproduction |
| Human TrkC Reporter Assay Kit |
1 x 96-well format assays |
IB27031 |
Human |
Growth Factor Receptor |
Cancer, Neurodegenerative |
| Human TrkC Reporter Assay Kit |
1 x 384-well format assays |
IB27032 |
Human |
Growth Factor Receptor |
Cancer, Neurodegenerative |
| Human VDR Reporter Assay Kit |
1 x 96-well format assays |
IB00701 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, Obesity, Osteoporosis, Reproduction |
| Human VDR Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB00701-32 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, Obesity, Osteoporosis, Reproduction |
| Human VDR Reporter Assay Kit |
1 x 384-well format assays |
IB00702 |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, Obesity, Osteoporosis, Reproduction |
| Human VEGFR Reporter Assay Kit |
1 x 96-well format assays |
IB15001 |
Human |
Growth Factor Receptor |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Kidney Disease, NASH/NAFLD, Neurodegenerative, Obesity, Reproduction |
| Human VEGFR Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB15001-32 |
Human |
Growth Factor Receptor |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Kidney Disease, NASH/NAFLD, Neurodegenerative, Obesity, Reproduction |
| Human VEGFR Reporter Assay Kit |
1 x 384-well format assays |
IB15002 |
Human |
Growth Factor Receptor |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Kidney Disease, NASH/NAFLD, Neurodegenerative, Obesity, Reproduction |
| in vitro Screening for Drug-Induced Hepatotoxicity Assay Kit |
2 x 48 assays in 96-well format |
ULC1003-48 |
Human |
|
|
| Mouse AhR Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
M06001 |
Mouse |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cancer, Cardiovascular, Dermatitis, Kidney Disease, NASH/NAFLD, Reproduction, Wound Healing |
| Mouse CAR Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
M00901 |
Mouse |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
NASH/NAFLD, Wound Healing |
| Mouse FXR Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
M00601 |
Mouse |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Reproduction |
| Mouse GR Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
M00201 |
Mouse |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Osteoporosis, Reproduction |
| Mouse LXRa Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
M00311 |
Mouse |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Mouse LXRa Reporter Assay Kit |
| Mouse LXRb Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
M00301 |
Mouse |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cancer, Cardiovascular, Dyslipidemia, Reproduction |
| Mouse PPARa Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
M00111 |
Mouse |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cancer, Cardiovascular, Dermatitis, Dyslipidemia, NASH/NAFLD, Obesity, Reproduction |
| Mouse PPARd Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
M00121 |
Mouse |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cancer, Dermatitis, Dyslipidemia, NASH/NAFLD, Neurodegenerative, Obesity, Reproduction, Wound Healing |
| Mouse PXR Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
M07001 |
Mouse |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cancer, Cardiovascular, Kidney Disease, NASH/NAFLD, Wound Healing |
| Mouse RORg Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
M04001 |
Mouse |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Autoimmune, Cancer, Dermatitis, NASH/NAFLD, Obesity |
| Mouse/Rat PPARg Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
MR00101 |
Rodent (Mouse / Rat Shared Ortholog) |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Osteoporosis, Reproduction |
| NFAT Reporter Assay Kit |
1 x 96-well format assays |
IB18001 |
Human |
Transcription Factor |
Cancer |
| NFAT Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB18001-32 |
Human |
Transcription Factor |
Cancer |
| NFAT Reporter Assay Kit |
1 x 384-well format assays |
IB18002 |
Human |
Transcription Factor |
Cancer |
| Panel of Human ER Reporter Assays: ERa & ERb |
Each in 1 x 48 96-well format |
IB00421-48P |
Human |
Nuclear Hormone Receptor |
|
| Panel of Human LXR Reporter Assays: LXRa & LXRb |
Each in 1 x 48 96-well format |
IB00321-48P |
Human |
Nuclear Hormone Receptor |
|
| Panel of Human PPAR Reporter Assays: PPARa, PPARd, & PPARg |
Each in 1 x 32 96-well format |
IB00131-32P |
Human |
Nuclear Hormone Receptor |
|
| Panel of Human RAR Reporter Assays: RARa, RARb & RARg |
Each in 1 x 32 96-well format |
IB02301-32P |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, Reproduction, Wound Healing |
| Panel of Human RXR Reporter Assays: RXRa, RXRb & RXRg |
Each in 1 x 32 96-well format |
IB00831-32P |
Human |
Nuclear Hormone Receptor |
Autoimmune, Cancer, Dermatitis, Reproduction, Wound Healing |
| Panel of Human TR Reporter Assays: TRa & TRb |
Each in 1 x 48 96-well format |
IB01201-48P |
Human |
Nuclear Hormone Receptor |
|
| Rabbit PGR Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
RB05001 |
Rabbit |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Obesity, Osteoporosis, Reproduction |
| Rat AhR Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
R06001 |
Rat |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cancer, Cardiovascular, Dermatitis, Kidney Disease, NASH/NAFLD, Reproduction, Wound Healing |
| Rat AR Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
R03001 |
Rat |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Autoimmune, Cancer, Cardiovascular, Dyslipidemia, Kidney Disease, NASH/NAFLD, Neurodegenerative, Obesity, Osteoporosis, Reproduction |
| Rat ERa Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
R00401 |
Rat |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Autoimmune, Cardiovascular, Dyslipidemia, NASH/NAFLD, Obesity, Osteoporosis, Reproduction, Wound Healing |
| Rat ERb Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
R00411 |
Rat |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Autoimmune, Cancer, Cardiovascular, Neurodegenerative, Obesity, Osteoporosis, Reproduction, Wound Healing |
| Rat FXR Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
R00601 |
Rat |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Reproduction |
| Rat GR Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
R00201 |
Rat |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Osteoporosis, Reproduction |
| Rat LXRb Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
R00301 |
Rat |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cancer, Cardiovascular, Dyslipidemia, Reproduction |
| Rat LXRα Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
R00311 |
Rat |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Autoimmune, Cancer, Cardiovascular, NASH/NAFLD, Reproduction |
| Rat PGR Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
R05001 |
Rat |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Obesity, Osteoporosis, Reproduction |
| Rat PPARa Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
R00111 |
Rat |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cancer, Cardiovascular, Dermatitis, Dyslipidemia, NASH/NAFLD, Obesity, Reproduction |
| Rat PPARd Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
R00121 |
Rat |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cancer, Dermatitis, Dyslipidemia, NASH/NAFLD, Neurodegenerative, Obesity, Reproduction, Wound Healing |
| Rat PXR Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
R07001 |
Rat |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cancer, Cardiovascular, Kidney Disease, NASH/NAFLD, Wound Healing |
| Rat RORγ Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
R04001 |
Rat |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Autoimmune, Cancer, Dermatitis, NASH/NAFLD, Obesity |
| TPOR Reporter Assay Kit |
1 x 96-well format assays |
IB20001 |
Human |
Cytokine Receptor |
Autoimmune, Reproduction |
| TPOR Reporter Assay Kit |
3 x 32 assays in 96-well format |
IB20001-32 |
Human |
Cytokine Receptor |
Autoimmune, Reproduction |
| TPOR Reporter Assay Kit |
1 x 384-well format assays |
IB20002 |
Human |
Cytokine Receptor |
Autoimmune, Reproduction |
| Zebrafish AhR Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
Z06001 |
Zebrafish |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cancer, Cardiovascular, Dermatitis, Kidney Disease, NASH/NAFLD, Reproduction, Wound Healing |
| Zebrafish AR Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
Z03001 |
Zebrafish |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Autoimmune, Cancer, Cardiovascular, Dyslipidemia, Kidney Disease, NASH/NAFLD, Neurodegenerative, Obesity, Osteoporosis, Reproduction |
| Zebrafish ERa Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
Z00401 |
Zebrafish |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Autoimmune, Cancer, Cardiovascular, Neurodegenerative, Obesity, Osteoporosis, Reproduction, Wound Healing |
| Zebrafish GR Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
Z00201 |
Zebrafish |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Osteoporosis, Reproduction |
| Zebrafish PPARg Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
Z00101 |
Zebrafish |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Autoimmune, Cancer, Cardiovascular, Dermatitis, Dyslipidemia, Kidney Disease, NASH/NAFLD, Obesity, Osteoporosis, Reproduction |
| Zebrafish RARa isoform A Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
Z02201 |
Zebrafish |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Autoimmune, Cancer, Dermatitis, Reproduction, Wound Healing |
| Zebrafish TRb Reporter Assay Kit |
1 x 96-well format assays / 3 x 32 assays in 96-well format |
Z01101 |
Zebrafish |
Nuclear Hormone Receptor Nuclear Receptor Orthologs |
Cancer, Cardiovascular, Dyslipidemia, NASH/NAFLD, Neurodegenerative, Obesity, Osteoporosis, Reproduction |