Universal Sequencing Technology (UST) was established by a group of NGS technology veterans from Roche/454, dedicated to the development of the most advanced cost-effective DNA sequencing technologies.
Short-read DNA sequencing platforms have dominated the NGS field ever since 2005 when Roche/454 Life Sciences launched the world’s first high throughput NGS sequencing platform. A well-known problem of short-read sequencing is that it cannot do de novo sequencing, and it does not provide the answers that are needed to have a full understanding of genome complexity and to connect health and traits to genome variations.
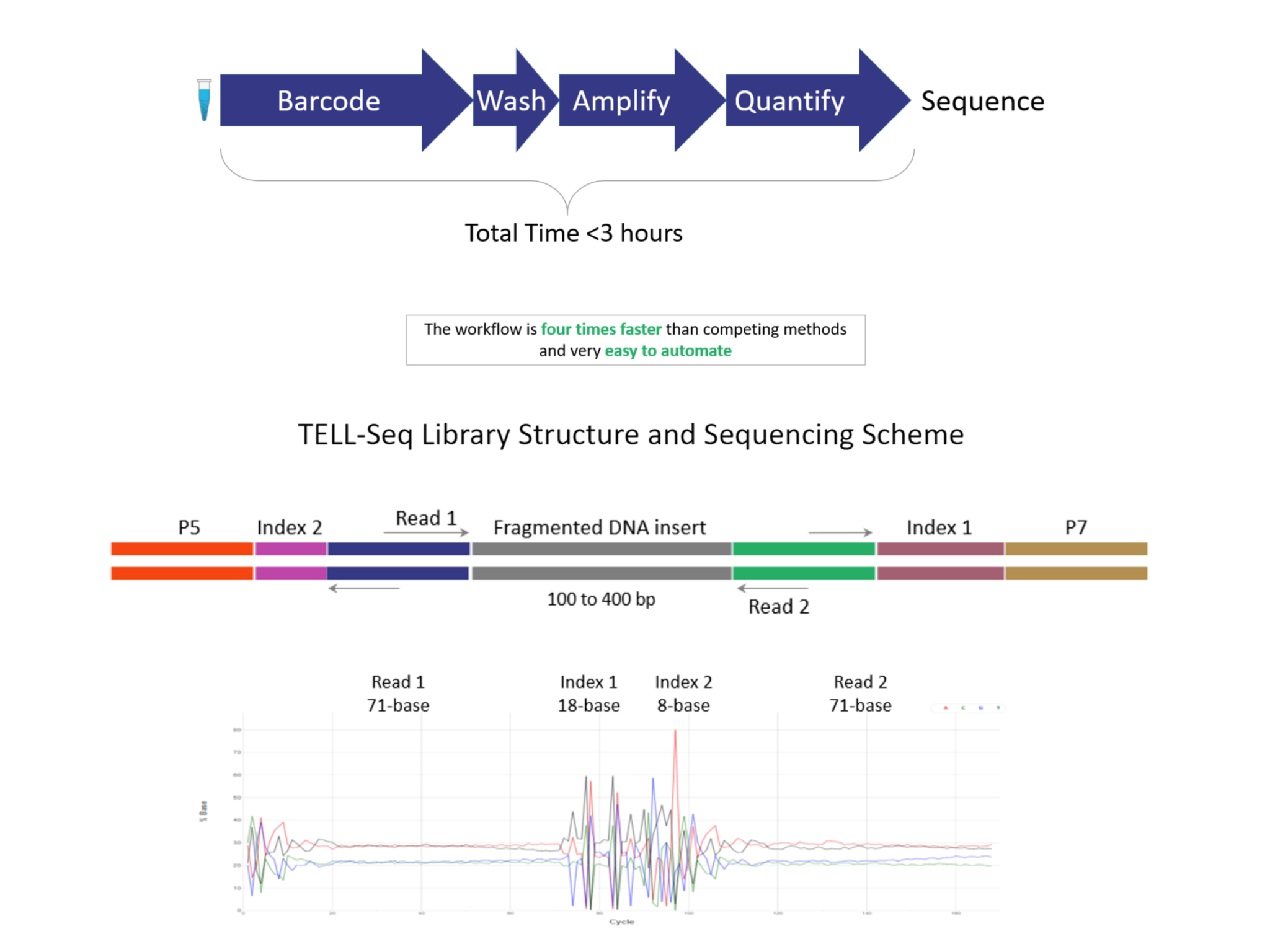
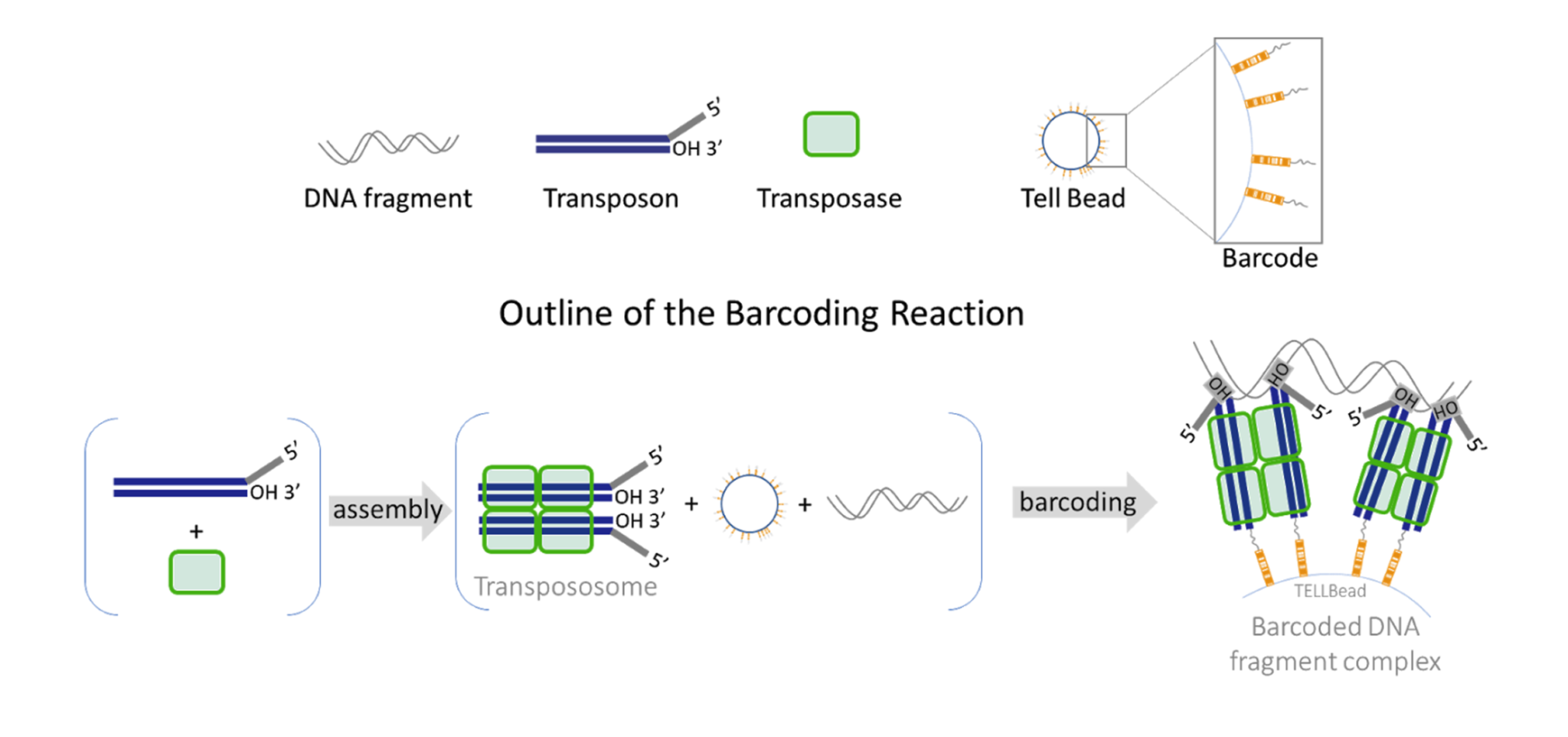
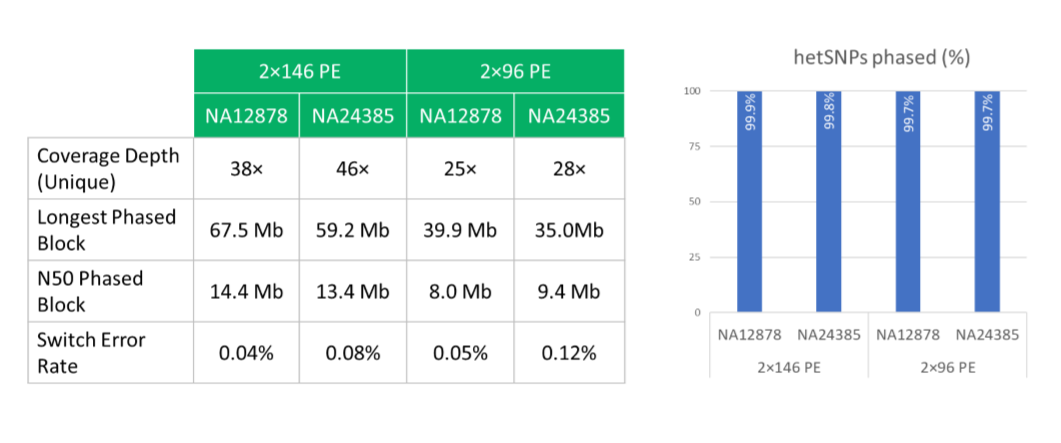
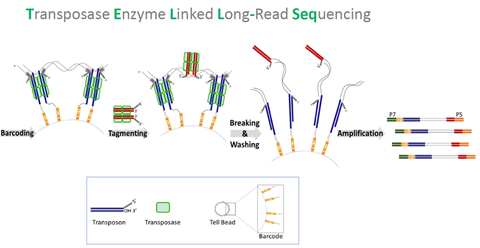
UST TELL-Seq enables short-read 2nd generation sequencing platforms to produce super long-read results (averaging at 40kb, but up to 200kb or longer). With UST TELL-Seq’s™ proven solution for library prep, a sequencing-ready library can be prepared in PCR tubes in only 3 hours and is highly accurate. As significantly cost-effective as UST TELL-Seq is, the technology requires very little DNA input (0.1-0.5ng for bacteria and target gene panels, 3-5ng for human genome).
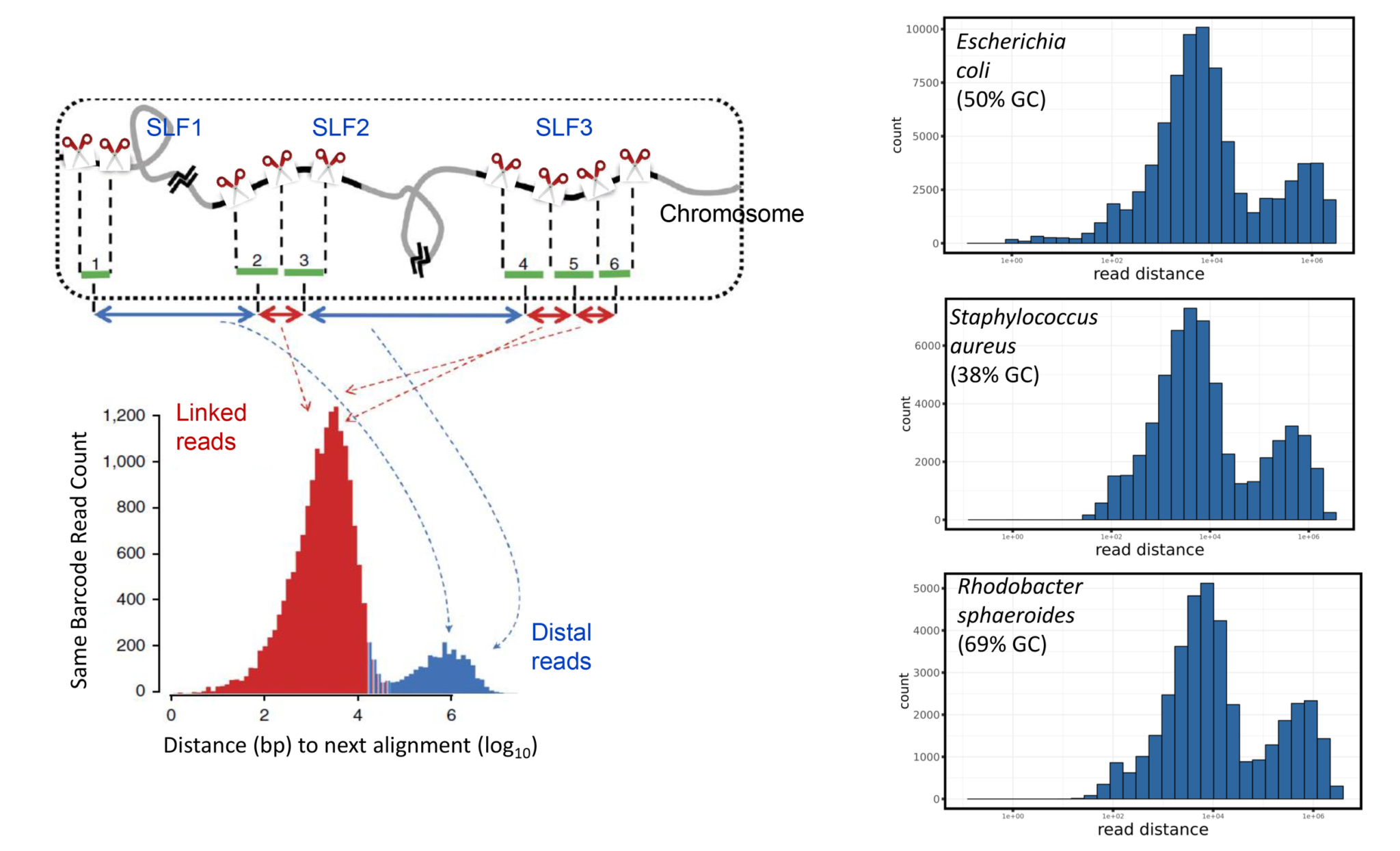
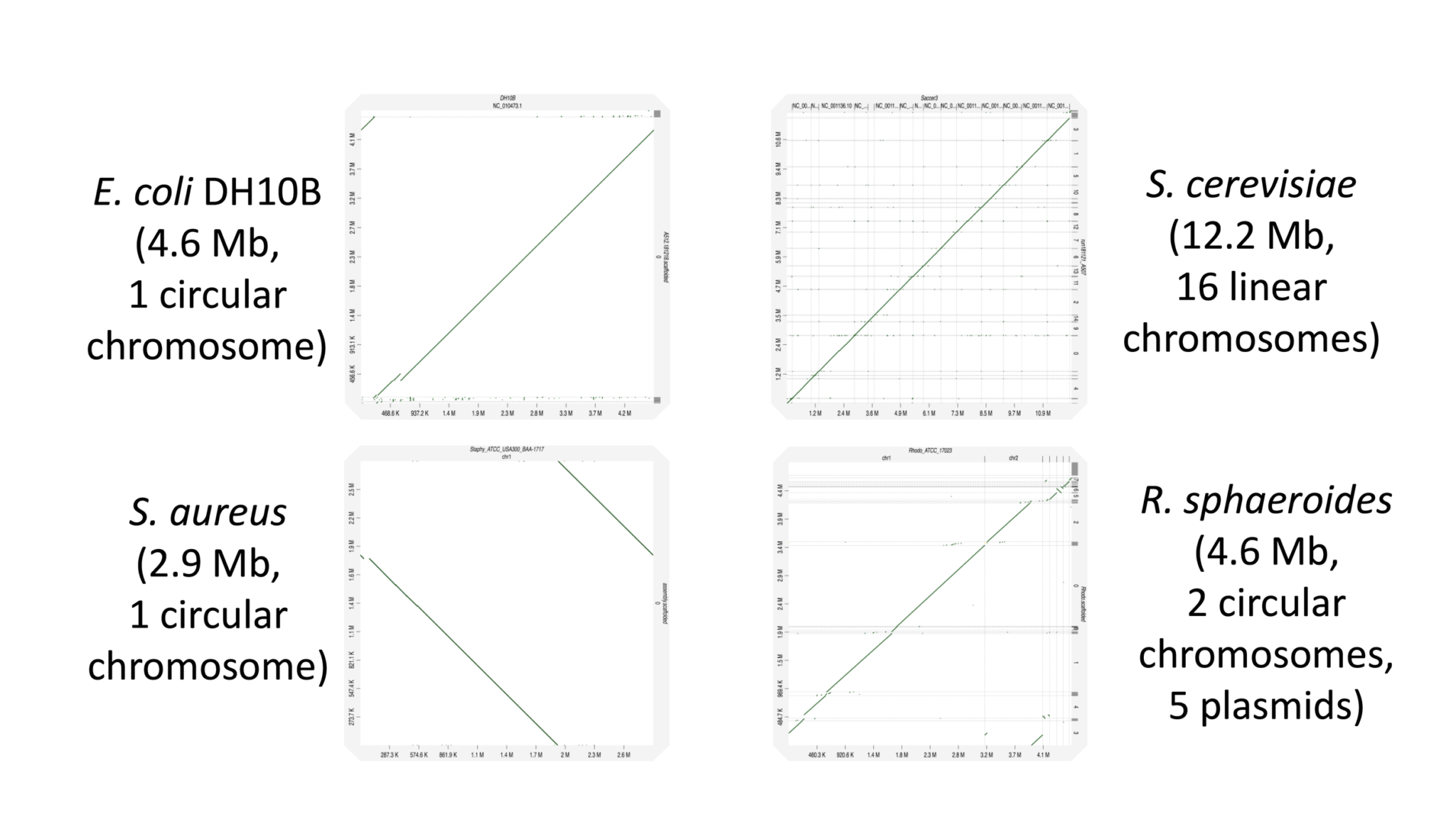
This technology allows researchers to do de novo sequencing (microbial, plants, animals, insects, etc.), and to identify structural variations among organisms and individuals more efficiently. Researchers can phase genomes to distinguish parental origins of structural variants. Phasing is particularly important in population genetic studies, newborn and adult health screening, and consumer genetics. In addition to phasing and structural variation analysis, the TELL-Seq kit can be used for metagenomics and analyzing complex mixtures of microbiomes in a wide variety of applications, such as medical microbiomes, environmental microbiomes, marine microbiomes, epidemical surveillance, hospital pathogen monitoring, and food safety.
In the process, Universal Sequencing Technology has filed more than 20 patent applications covering linked read and single-cell library preparation chemistry and groundbreaking 3rd generation DNA sequencing technology.
Universal Sequencing will lead the next wave of DNA sequencing innovations.