Precision medicine, also known as personalized or individualized medicine, describes the use of patient-specific molecular features to guide disease diagnosis, monitor progression and treatment response, or identify new targets for drug development. The application of precision medicine principles enabled strides in cancer therapeutics and is poised to significantly alter the treatment of multiple pathologies, including metabolic & cardiovascular disease, autoimmune & inflammatory disease, and neurodevelopmental & neurodegenerative disease. Here, we will review the foundational ideas and challenges underlying precision medicine and discuss how the epigenome and chromatin biomarkers may be key to advancing this important research.
The promise of chromatin research to advance precision medicine
In this blog:
The need for precision medicine
Historically, most therapeutics have been utilized across broad patient populations. However, disease etiology is complex, often arising from an interplay between multiple genetic, environmental, and lifestyle factors.
Consequently, therapeutic outcomes can vary widely, from success – in the form of total remission or relief of symptoms – to moderate or severe failure – including increased side effects or no change in disease progression. Precision medicine is a potential solution to this problem (1,2). The overarching goal of precision medicine is to use patient-specific molecular features to inform:
- Diagnosis. What characterizes the disease?
- Patient stratification. Can we use biomarkers to refine subtypes of complex disease?
- Prognosis. How severe is the disease? How rapidly will it progress?
- Treatment selection. What is the optimal course of treatment for this specific patient?
- Treatment response indicators. Is the drug working as expected or should we change dosage, try another treatment, or supplement with another drug?
- Novel drug development. Can we identify new therapeutic targets?
In pursuit of these answers, many research dollars have been spent on efforts to identify suitable therapeutic targets and biomarkers. Below, we highlight current approaches, clinical successes, and future opportunities for precision medicine research.
Founding principles of precision medicine
Genetic variant-based approaches
While the concept of precision medicine has existed since the 1960s, the movement gained real momentum following the completion of the human genome project in the early 2000s1. This feat enabled scientists to conduct large studies of human genetic variants (e.g. mutations and single nucleotide polymorphisms) and identify links to unique phenotypes or diseases (3). These projects, called genome-wide association studies (GWAS), identified hundreds of thousands of risk variants for diseases and traits (4).
In some instances, the identification of disease-associated risk variants enlightened underlying disease biology, which in turn drove therapeutic development. In other cases, GWAS identified care-informing biomarkers that are used to select patient-appropriate treatments. Below we highlight a few key applications:
Case Study 1: Crohn’s disease and IL-12/IL-23 inhibitors
In Crohn’s disease, GWAS implicated cytokines IL-12 and IL-23 in disease development (5). Basic research into the pro-inflammatory effects of these molecules resulted in successful immunotherapeutic drug trials targeting this pathway (6).
Case Study 2: Lung cancer biomarkers for treatment selection
DNA sequencing efforts have uncovered multiple lung cancer-associated genetic variants (7), seven of which have FDA-approved therapeutics (8). This allows physicians to use genetic testing to select treatments and (hopefully) improve patient outcomes.
However, the identification of disease-associated risk variants alone may provide an incomplete view of disease etiology. Thus, to fully leverage these findings for precision medicine, it is essential to understand their functional consequences. For example, variants in protein coding regions of the genome could impact protein structure, function, and/or expression. Each represents a distinct molecular outcome that may need to be treated differently, as highlighted by cystic fibrosis.
Case Study 3: Genetic risk variants inform cystic fibrosis treatment
Multiple genetic variants are associated with cystic fibrosis. Depending on the causal variant, two drugs with distinct mechanisms of action on the CFTR protein – aiding in trafficking to the cell surface or ion channel function – may be used alone or in combination (9-11).
Notably, more than 90% of GWAS-identified variants are in noncoding regions of the genome (12), which is enriched for cis-regulatory elements (i.e. enhancers) that activate gene expression in a cell-type specific manner (13). As a result, many researchers focused on how disease risk variants impact transcriptional regulation as opposed to protein structure or function (14-16). The ongoing challenge lies in knowing what the target gene(s) are. Noncoding regions can act through direct and indirect mechanisms across large genomic distances; therefore, the gene closest to the disease variant may not always be the causal determinant1 (7,18).
Learn the basics of chromatin mapping!
Gene expression-based approaches
Researchers have found substantial changes in gene expression across disease states, including cancer, autoimmune disorders, neurodegenerative disease, cardiovascular disease, and others (19-23). This highlights the necessity of genome-wide survey approaches to uncover transcriptional signatures in disease. To this end, gene expression studies, including microarray and RNA-sequencing approaches, have helped identify subtypes of many diseases.
Case Study 4: Breast cancer patient stratification and personalized therapy selection
Thanks in part to the application of precision medicine-based diagnostics and treatments, the five-year survival rate of breast cancer patients has increased dramatically (24) (from 76% in 1975 to now >90% (25)). Breast cancer can be categorized by the expression of multiple signaling receptors to inform diagnosis, selection of the appropriate therapeutic combination, and recurrence risk. For example, patients positive for the expression of the tyrosine kinase HER2 (HER2+) often receive trastuzumab, a monoclonal antibody that targets HER2 (26). Integration of molecular classifications such as HER2 status with quantitative measurements of cell differentiation and proliferation also inform patient prognosis (27). These are just a few examples of the vast findings that pushed the field forward in helping physicians treat and support breast cancer patients and their families.
Research in gene expression-informed personalized medicine remains ongoing, both to further refine subclasses of cancers with established biomarkers, such as breast cancer (28), and those without, such as some brain cancers (29). These findings will yield novel biomarker and therapeutic targets, potentially saving many lives.
Shortcomings of current precision medicine approaches
Above we highlighted clinical successes in many aspects of precision medicine. Despite these remarkable achievements, GWAS and transcriptomics often fail to generate clinically relevant biomarkers or drug targets, leaving many diseases difficult to diagnose and treat. This can be attributed to many outstanding questions:
Which variant is actually responsible for the disease?
These mutations rarely occur in isolation. In fact, variants are typically inherited as sets in large blocks of DNA called haplotypes, making it difficult to know which (if any) could be influencing disease state.
How do genetic and environmental influences intersect?
Indeed, the penetrance of some genetic variants is modulated by lifestyle30 or environmental factors (31), such as use of certain prescription drugs, exercise, and pollution, where their gene regulatory influence and disease risk are correlated with exposure.
Are environmental factors more likely the cause?
Some disease-associated changes in gene regulation are not associated with genetic variants at all. Instead, the root cause of these gene regulatory changes – associated with states ranging from allergies (32) to low birth weight (33) to heart disease (34) – is dependent on environmental exposure.
Is gene expression capturing everything we need to know?
Current approaches studying aberrant RNA expression provide a snapshot of what is currently happening inside the cell. They cannot provide information regarding what a cell is primed to do, such as treatment-induced changes in genetic programs or development of drug resistance. This lack of mechanistic information leaves many open questions regarding treatment response and potential for relapse that are hugely informative to personalized care.
Why are we only focusing on protein-coding regions?
Gene expression and genetic variants are major indicators of disease – but they are only part of the picture. In fact, less than 1% of the human genome is protein coding (35). Gene regulation is primarily controlled by noncoding regions, which can act as enhancers, form chromatin loops, bind transcription factors, or recruit epigenetic modifying enzymes & reader proteins. In addition, many disease risk variants are also enriched in noncoding DNA. Thus, noncoding DNA and chromatin-based mechanisms represent a massive untapped pool of biomarkers and drug targets.
The convergence of genetics, environment, transcription, and chromatin structure on the development of disease phenotypes is complex (Figure 1). Current approaches generally fail to account for these diverse factors, making it difficult to predict disease progression and response to treatment.
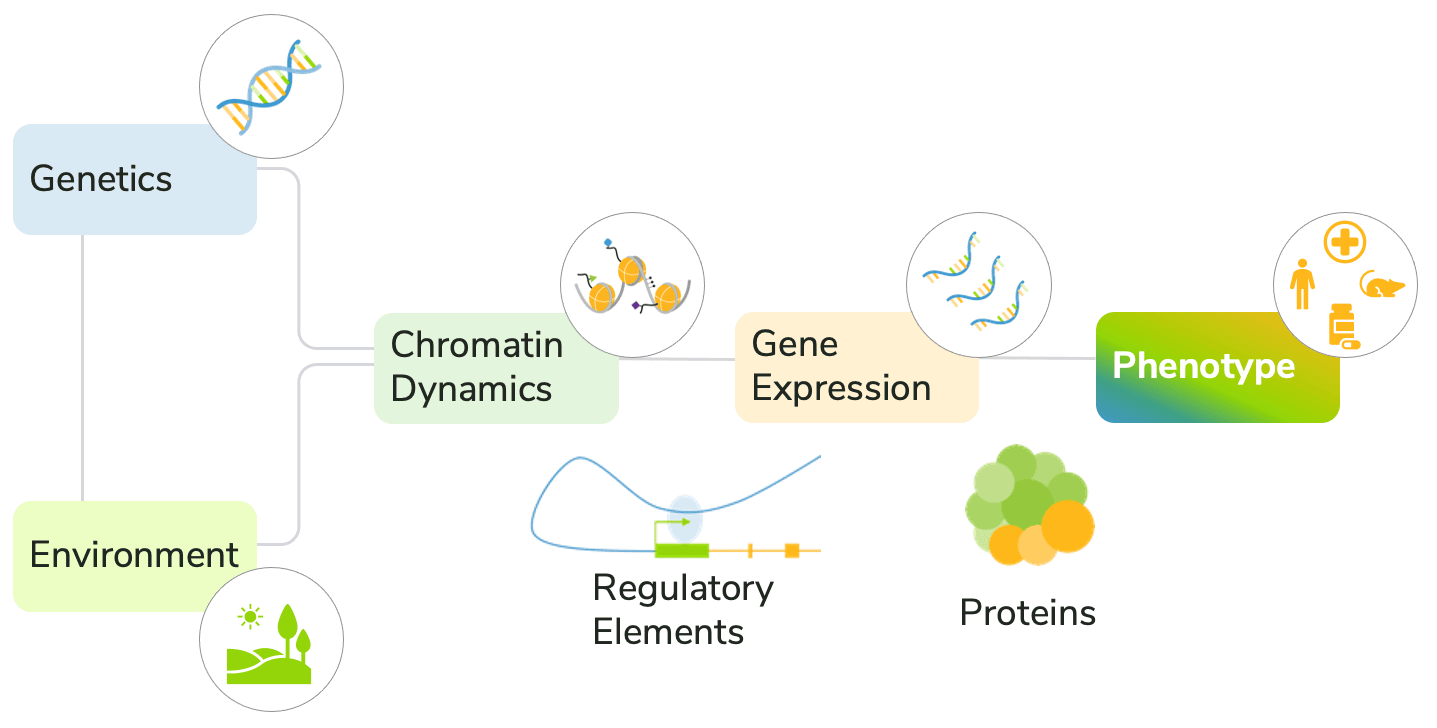
Figure 1: Environmental and molecular factors together drive disease development.
Integration of epigenomics into precision medicine
The epigenome could hold the key to identifying novel biomarkers and drug targets to advance precision medicine. The epigenome comprises a complex molecular code that regulates chromatin structure and function to control gene expression. As such, studying the epigenome could serve as an important molecular tool to help us understand changes in gene expression resulting from genetic variants, the environment, and therapeutics.
To that end, chromatin modifications, including DNA methylation and histone post-translational modifications (PTMs), have provided remarkable insight into gene regulatory programs and are becoming an essential aspect of precision medicine research.
DNA methylation
Historically, mapping DNA methylation has been the preferred approach for epigenomic precision medicine research. This established repressive chromatin mark has multiple options for detection: methylation-sensitive PCR & restriction enzymes, antibody-based methods (e.g. MeDIP), or bisulfite conversion followed by sequencing (BS-seq/WGBS) (36,37). The integration of DNA methylation into diagnostics has resulted in exciting advances:
Case Study 5: Classification and treatment selection for central nervous system (CNS) tumors
Many primary CNS tumors are difficult to classify by conventional histological methods (38). For example, CNS neuroectodermal tumors are a broad class histologically characterized by small, poorly differentiated neurons and glia. In one study, the integration of DNA methylation profiling reclassified over 75% of tumors to more specific subtypes such as medulloblastomas, high-grade gliomas, ependymomas, and pineal tumors (39). In other primary CNS tumors, such as glioblastoma, the methylation of select promoters (e.g. MGMT) promotes sensitivity to treatments, such as temozolomide, guiding therapeutic selection (40).
Case Study 6: Non-invasive in vitro diagnostic development
DNA methylation was detected on circulating cell-free nucleosomes in patient plasma/serum in early studies and has been observed in several proof-of-concept clinical studies, such as lung, breast, and colorectal cancers (37,41-43). This paved the way for development of FDA-approved in vitro diagnostic tests from easily obtained samples, such as stool or blood for Cologuard® and Epi proColon®, respectively, as well as several promising liquid biopsy tests, including GRAIL’s Galleri™ to detect cancer regardless of its type (44-46).
However, due to the inability to provide granular details about distinct chromatin compartments, particularly tissue/cell-specific enhancer activity, DNA methylation is not sufficient for some applications. Furthermore, DNA methylation is not always a repressive mark (47), meaning its impact on gene expression can be ambiguous. While DNA methylation has proven that deciphering the epigenome can inform precision medicine approaches, it is only the tip of the iceberg in terms of unraveling epigenomics and gene expression-modifying mechanisms on chromatin.
Histone post-translational modifications
The remarkable chemical and protein diversity on chromatin is brimming with potential precision medicine markers. Specifically, histone PTMs and chromatin-bound proteins (such as transcription factors) exert powerful and dynamic influences on gene expression. To date, over 100 unique histone PTMs or combinations thereof have been linked to human disease, including multiple cancers (48-53), and global alterations to histone PTM patterns have been found to be predictive of disease state, recurrence, and patient response (54-56).
Unlike DNA methylation or even chromatin accessibility assays (e.g. ATAC-seq), histone PTMs provide insight into distinct, specific genomic features. The development of genome-wide chromatin mapping tools helped assign histone PTMs to specific genomic compartments (Figure 2), such as active enhancers (H3K27ac), active promoters (H3K4me3), active gene bodies (H3K36me3), and even repressed genes (H3K9me2/3, H3K27me3). Therefore, studying histone PTMs can illuminate the diversity of the gene regulatory landscape – including what a cell is primed to do. A classic example is the bivalent H3K4me3/H3K27me3 mark, which denotes enhancers and promoters that are “poised” for activation in undifferentiated stem/progenitor cells57. Notably, altered expression of bivalent genes is frequently observed in cancer58, where they can be associated with more severe progression (59) and/or drug resistance (60).
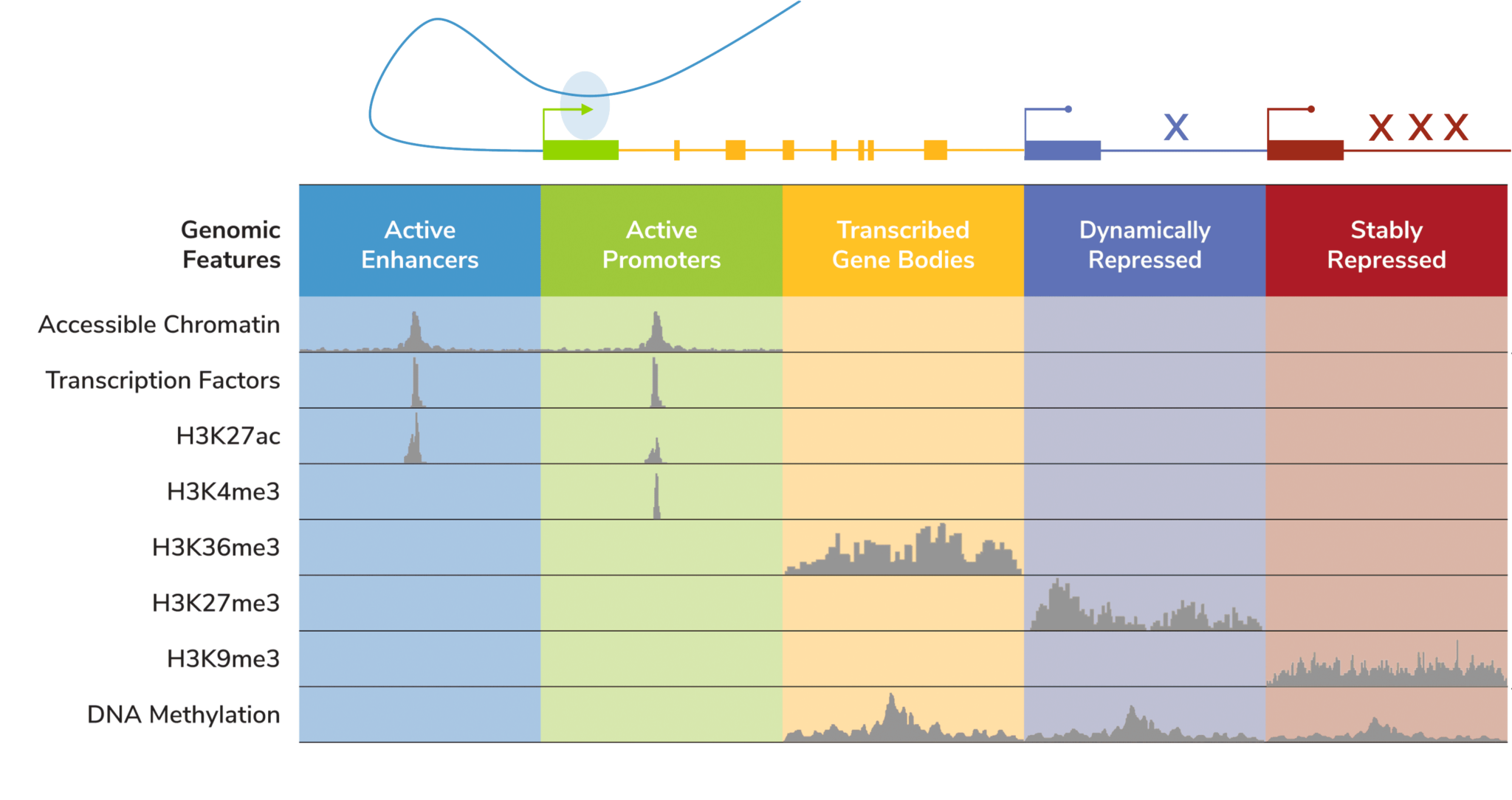
Figure 2: Overview of the localization of chromatin features.
As such, genomic mapping studies contain crucial information for personalized medicine. Cutting-edge research in this area is already making an impact in the precision medicine space:
Case Study 7: Prediction and regulation of treatment response in cancer
One study found the histone PTM H3K27ac, a marker of enhancers and active transcriptional programs, could be used to distinguish sub-types of colorectal cancer, including prognostic outcomes and responses to treatment (61). Further, a novel combination therapy using these enhancer sub-type classifications showed efficacy in patient-derived xenografts. Another study found that the repressive histone PTM H3K27me3 sensitizes breast cancer cells to chemotherapy, while preventing this mark inhibits drug tolerance and delays tumor formation (60), pointing to the therapeutic potential of histone PTMs.
Together, these studies demonstrate how epigenomic studies have strong potential to guide clinical decision making and drug development.
Learn about CUT&RUN and CUT&Tag!
Challenges associated with integrating epigenomics
While chromatin mapping shows great promise for precision medicine applications, the epigenome has been challenging to study in the context of clinical applications.
Historically, chromatin mapping was conducted via ChIP-seq. While groundbreaking, it has significant limitations that have hindered its use in precision medicine. Specifically, these include:
- Low sensitivity and throughput, requiring millions of cells, deep sequencing, and extensive hands-on time.
- Difficulty in adaptation across cell types, requiring extensive optimization for each new cell type.
- Nonspecific antibodies to histone PTMs – in fact, the vast majority (>70% as shown by EpiCypher62) cross-react with related marks
- All of these result in high costs and low-quality data!
Together, these challenges limit the scale, application, and interpretation of ChIP-seq data. The poor reliability of ChIP has also made it difficult to study disease heterogeneity and patient-specific treatment response.
In sum, ChIP-seq is incapable of doing what epigenomics needs to do for precision medicine. What other options are there?
CUT&RUN and CUT&Tag: improved epigenomics for precision medicine
CUT&RUN/CUT&Tag assays address many of the problems associated with ChIP-seq, generating high resolution profiles with a fraction of the cells and sequencing reads. It’s also fast – EpiCypher’s CUTANA™ CUT&RUN/CUT&Tag assays can go from cells to sequencing libraries in just two to three days, compared to a week (or more) for ChIP.
CUT&RUN and CUT&Tag are ideal for these studies and could advance our knowledge of disease risk variants, gene regulatory programs, and diagnostic/therapeutic biomarkers. Advantages and features relevant for personalized medicine applications as compared to ChIP-seq include:
- High sensitivity and throughput: streamlined workflows and kits requiring as few as 5,000 cells
- Broad cell type compatibility: stem cells, immune cells, patient-derived xenografts, FACS-sorted cells, and mouse and human primary cells, frozen samples, and lightly cross-linked materials
- In-assay validated antibodies, to ensure confidence in your results.
- Low costs – approximately 70% less per reaction compared to ChIP-seq
CUT&RUN is robust for most targets, including transiently interacting proteins (e.g., chromatin remodeling enzymes), pioneer factors, heterochromatin targets and has single-cell compatible protocols (63), enabling analysis of heterogeneous tissues. Importantly, CUT&Tag is only recommended for histone PTM targets.
Many groups have already used EpiCypher’s CUT&RUN (64-67) and CUT&Tag (68,69) assays to study chromatin structure for biomedical research. Other publications have already demonstrated the promise of this technology for precision medicine:
Case Study 8: Disease-associated enhancer identification in neurons
By combining H3K27ac CUT&RUN, ATAC-seq, and RNA-seq, one group was able to identify enhancers unique to excitatory vs. inhibitory human neurons (70). Enhancer maps were integrated with a database of psychiatric disease risk variants, which revealed enrichment connected to schizophrenia, ADHD, and bipolar disorder. This study directly demonstrates how integration of epigenomics with existing datasets can further the understanding of disease risk variant function and aberrant gene expression to drive precision medicine.
Case Study 9: Identification of transcriptional footprints for liquid biopsy
CUT&RUN has been used to identify ER+ breast cancer transcriptional footprints in xenografts (71). The transcriptional profiles were used to subclassify human breast tumors and, remarkably, were detected in patient plasma (71). These studies directly demonstrate the potential of next-generation CUT&RUN genomic mapping technologies to advance emerging non-invasive liquid biopsy personalized medicine applications
Conclusions and future perspectives
Precision medicine is an exciting field challenged by the difficulty of defining patient-specific molecular features. The study of chromatin for precision medicine has the potential to significantly alter the field in two main ways:
- Identification and validation of novel chromatin biomarkers – representing an untapped area of regulatory and environmentally-sensitive diversity over existing genomic and transcriptomic approaches.
- Elucidation of transcriptional regulatory mechanisms driving disease, which will help characterize disease risk variants and identify new transcriptional biomarkers and drug targets.
CUT&RUN/CUT&Tag are powerful assays that can be leveraged to achieve these goals. Additional applications of epigenomics in precision medicine include fine mapping of chromatin compartments in unique cell types to help elucidate chromatin regulatory mechanisms underlying disease development and progression. Such epigenomic maps can be integrated with other sequencing modalities, including DNA methylation, RNA-seq, ATAC-seq, or Hi-C, to characterize diseases across individual patients and improve clinical outcomes.
- Jørgensen JT. Twenty Years with Personalized Medicine: Past, Present, and Future of Individualized Pharmacotherapy. Oncologist 24, e432-e40 (2019). PubMed PMID: 30940745.
- Ashley EA. Towards precision medicine. Nat Rev Genet 17, 507-22 (2016). PubMed PMID: 27528417.
- Ikegawa S. A short history of the genome-wide association study: where we were and where we are going. Genomics Inform 10, 220-5 (2012). PubMed PMID: 23346033.
- Uffelmann E et al. Genome-wide association studies. Nature Reviews Methods Primers 1, 59 (2021). https://doi.org/10.1038/s43586-021-00056-9.
- Wang K et al. Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn Disease. Am J Hum Genet 84, 399-405 (2009). PubMed PMID: 19249008.
- Moschen AR et al. IL-12, IL-23 and IL-17 in IBD: immunobiology and therapeutic targeting. Nat Rev Gastroenterol Hepatol 16, 185-96 (2019). PubMed PMID: 30478416.
- Greulich H. The genomics of lung adenocarcinoma: opportunities for targeted therapies. Genes Cancer 1, 1200-10 (2010). PubMed PMID: 21779443.
- Targeted Therapies for Lung Cancer: American Lung Association. Available from: https://www.lung.org/lung-health-diseases/lung-disease-lookup/lung-cancer/treatment/types-of-treatment/targeted-therapies.
- Boyle MP et al. A CFTR corrector (lumacaftor) and a CFTR potentiator (ivacaftor) for treatment of patients with cystic fibrosis who have a phe508del CFTR mutation: a phase 2 randomised controlled trial. Lancet Respir Med 2, 527-38 (2014). PubMed PMID: 24973281.
- Condren ME et al. Ivacaftor: a novel gene-based therapeutic approach for cystic fibrosis. J Pediatr Pharmacol Ther 18, 8-13 (2013). PubMed PMID: 23616732.
- Wainwright CE et al. Lumacaftor-Ivacaftor in Patients with Cystic Fibrosis Homozygous for Phe508del CFTR. N Engl J Med 373, 1783-4 (2015). PubMed PMID: 26510034.
- Edwards SL et al. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet 93, 779-97 (2013). PubMed PMID: 24210251.
- Gasperini M et al. Towards a comprehensive catalogue of validated and target-linked human enhancers. Nat Rev Genet 21, 292-310 (2020). PubMed PMID: 31988385.
- Morley M et al. Genetic analysis of genome-wide variation in human gene expression. Nature 430, 743-7 (2004). PubMed PMID: 15269782.
- Schadt EE et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet 37, 710-7 (2005). PubMed PMID: 15965475.
- Emilsson V et al. Genetics of gene expression and its effect on disease. Nature 452, 423-8 (2008). PubMed PMID: 18344981.
- McGovern A et al. Capture Hi-C identifies a novel causal gene, IL20RA, in the pan-autoimmune genetic susceptibility region 6q23. Genome Biol 17, 212 (2016). PubMed PMID: 27799070.
- Smemo S et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507, 371-5 (2014). PubMed PMID: 24646999.
- Bradner JE et al. Transcriptional Addiction in Cancer. Cell 168, 629-43 (2017). PubMed PMID: 28187285.
- Nagafuchi Y et al. Lessons From Transcriptome Analysis of Autoimmune Diseases. Front Immunol 13, 857269 (2022). PubMed PMID: 35663941.
- Cooper-Knock J et al. Gene expression profiling in human neurodegenerative disease. Nat Rev Neurol 8, 518-30 (2012). PubMed PMID: 22890216.
- Liu Y et al. RNA-Seq identifies novel myocardial gene expression signatures of heart failure. Genomics 105, 83-9 (2015). PubMed PMID: 25528681.
- Lee TI et al. Transcriptional regulation and its misregulation in disease. Cell 152, 1237-51 (2013). PubMed PMID: 23498934.
- Siegel RL et al. Cancer statistics, 2022. CA Cancer J Clin 72, 7-33 (2022). PubMed PMID: 35020204.
- Cancer Stat Facts: Female Breast Cancer: National Cancer Institute. Available from: https://seer.cancer.gov/statfacts/html/breast.html.
- Slamon DJ et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 344, 783-92 (2001). PubMed PMID: 11248153.
- Sotiriou C et al. Gene-expression signatures in breast cancer. N Engl J Med 360, 790-800 (2009). PubMed PMID: 19228622.
- Zhao W et al. SCD5 expression correlates with prognosis and response to neoadjuvant chemotherapy in breast cancer. Sci Rep 11, 8976 (2021). PubMed PMID: 33903614.
- Yuan Q et al. Identifying Differential Expression Genes and Prognostic Signature Based on Subventricular Zone Involved Glioblastoma. Front Genet 13, 912227 (2022). PubMed PMID: 35873494.
- Knowles DA et al. Allele-specific expression reveals interactions between genetic variation and environment. Nat Methods 14, 699-702 (2017). PubMed PMID: 28530654.
- Favé MJ et al. Gene-by-environment interactions in urban populations modulate risk phenotypes. Nat Commun 9, 827 (2018). PubMed PMID: 29511166.
- Prescott S et al. The role of epigenetic dysregulation in the epidemic of allergic disease. Clin Epigenetics 2, 223-32 (2011). PubMed PMID: 21949548.
- Heijmans BT et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A 105, 17046-9 (2008). PubMed PMID: 18955703.
- Breitling LP et al. Smoking, F2RL3 methylation, and prognosis in stable coronary heart disease. Eur Heart J 33, 2841-8 (2012). PubMed PMID: 22511653.
- Sheffield NC et al. Identifying and characterizing regulatory sequences in the human genome with chromatin accessibility assays. Genes (Basel) 3, 651-70 (2012). PubMed PMID: 24705081.
- Mattei AL et al. DNA methylation: a historical perspective. Trends Genet 38, 676-707 (2022). PubMed PMID: 35504755.
- Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer 3, 253-66 (2003). PubMed PMID: 12671664.
- Galbraith K et al. DNA methylation as a diagnostic tool. Acta Neuropathol Commun 10, 71 (2022). PubMed PMID: 35527288.
- Sturm D et al. New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs. Cell 164, 1060-72 (2016). PubMed PMID: 26919435.
- Butler M et al. MGMT Status as a Clinical Biomarker in Glioblastoma. Trends Cancer 6, 380-91 (2020). PubMed PMID: 32348734.
- Li P et al. Liquid biopsies based on DNA methylation as biomarkers for the detection and prognosis of lung cancer. Clin Epigenetics 14, 118 (2022). PubMed PMID: 36153611.
- Manoochehri M et al. DNA methylation biomarkers for noninvasive detection of triple-negative breast cancer using liquid biopsy. Int J Cancer 152, 1025-35 (2023). PubMed PMID: 36305646.
- Jin S et al. Efficient detection and post-surgical monitoring of colon cancer with a multi-marker DNA methylation liquid biopsy. Proc Natl Acad Sci U S A 118, (2021). PubMed PMID: 33495330.
- Pickhardt PJ. Emerging stool-based and blood-based non-invasive DNA tests for colorectal cancer screening: the importance of cancer prevention in addition to cancer detection. Abdom Radiol (NY) 41, 1441-4 (2016). PubMed PMID: 27259335.
- Nadauld LD et al. The PATHFINDER Study: Assessment of the Implementation of an Investigational Multi-Cancer Early Detection Test into Clinical Practice. Cancers (Basel) 13, (2021). PubMed PMID: 34298717.
- Taryma-Leśniak O et al. Current status of development of methylation biomarkers for in vitro diagnostic IVD applications. Clin Epigenetics 12, 100 (2020). PubMed PMID: 32631437.
- Rauluseviciute I et al. DNA hypermethylation associated with upregulated gene expression in prostate cancer demonstrates the diversity of epigenetic regulation. BMC Med Genomics 13, 6 (2020). PubMed PMID: 31914996.
- Chopra M et al. Disturbing the histone code in leukemia: translocations and mutations affecting histone methyl transferases. Cancer Genet 208, 192-205 (2015). PubMed PMID: 25592767.
- Greenblatt SM et al. Chromatin modifiers and the promise of epigenetic therapy in acute leukemia. Leukemia 28, 1396-406 (2014). PubMed PMID: 24609046.
- Gajer JM et al. Histone acetyltransferase inhibitors block neuroblastoma cell growth in vivo. Oncogenesis 4, e137 (2015). PubMed PMID: 25664930.
- Witt O et al. Targeting histone deacetylases in neuroblastoma. Curr Pharm Des 15, 436-47 (2009). PubMed PMID: 19199971.
- Hanmod SS et al. Targeting histone deacetylases (HDACs) and Wee1 for treating high-risk neuroblastoma. Pediatr Blood Cancer 62, 52-9 (2015). PubMed PMID: 25308916.
- Kobayashi K et al. Epigenetic regulation of the neuroblastoma genes, Arid3b and Mycn. Oncogene 32, 2640-8 (2013). PubMed PMID: 22751132.
- Seligson DB et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 435, 1262-6 (2005). PubMed PMID: 15988529.
- Kamińska K et al. Prognostic and Predictive Epigenetic Biomarkers in Oncology. Mol Diagn Ther 23, 83-95 (2019). PubMed PMID: 30523565.
- Seligson DB et al. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol 174, 1619-28 (2009). PubMed PMID: 19349354.
- Macrae TA et al. Regulation, functions and transmission of bivalent chromatin during mammalian development. Nat Rev Mol Cell Biol 24, 6-26 (2023). PubMed PMID: 36028557.
- Kumar D et al. Decoding the function of bivalent chromatin in development and cancer. Genome Res 31, 2170-84 (2021). PubMed PMID: 34667120.
- Chaffer CL et al. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 154, 61-74 (2013). PubMed PMID: 23827675.
- Marsolier J et al. H3K27me3 conditions chemotolerance in triple-negative breast cancer. Nat Genet 54, 459-68 (2022). PubMed PMID: 35410383.
- Orouji E et al. Chromatin state dynamics confers specific therapeutic strategies in enhancer subtypes of colorectal cancer. Gut 71, 938-49 (2022). PubMed PMID: 34059508.
- Shah RN et al. Examining the Roles of H3K4 Methylation States with Systematically Characterized Antibodies. Mol Cell 72, 162-77.e7 (2018). PubMed PMID: 30244833.
- Hainer SJ et al. Profiling of Pluripotency Factors in Single Cells and Early Embryos. Cell 177, 1319-29.e11 (2019). PubMed PMID: 30955888.
- Janssens DH et al. Automated in situ chromatin profiling efficiently resolves cell types and gene regulatory programs. Epigenetics Chromatin 11, 74 (2018). PubMed PMID: 30577869.
- Theisen ER et al. Chromatin profiling reveals relocalization of lysine-specific demethylase 1 by an oncogenic fusion protein. Epigenetics 16, 405-24 (2021). PubMed PMID: 32842875.
- Garcia-Martinez L et al. Endocrine resistance and breast cancer plasticity are controlled by CoREST. Nat Struct Mol Biol 29, 1122-35 (2022). PubMed PMID: 36344844.
- Dai X et al. Massively parallel knock-in engineering of human T cells. Nat Biotechnol (2023). PubMed PMID: 36702900.
- Sparbier CE et al. Targeting Menin disrupts the KMT2A/B and polycomb balance to paradoxically activate bivalent genes. Nat Cell Biol 25, 258-72 (2023). PubMed PMID: 36635503.
- Battistello E et al. Stepwise activities of mSWI/SNF family chromatin remodeling complexes direct T cell activation and exhaustion. Mol Cell 83, 1216-36.e12 (2023). PubMed PMID: 36944333.
- Sanchez-Priego C et al. Mapping cis-regulatory elements in human neurons links psychiatric disease heritability and activity-regulated transcriptional programs. Cell Rep 39, 110877 (2022). PubMed PMID: 35649373.
- Rao S et al. Transcription factor-nucleosome dynamics from plasma cfDNA identifies ER-driven states in breast cancer. Sci Adv 8, eabm4358 (2022). PubMed PMID: 36001652.