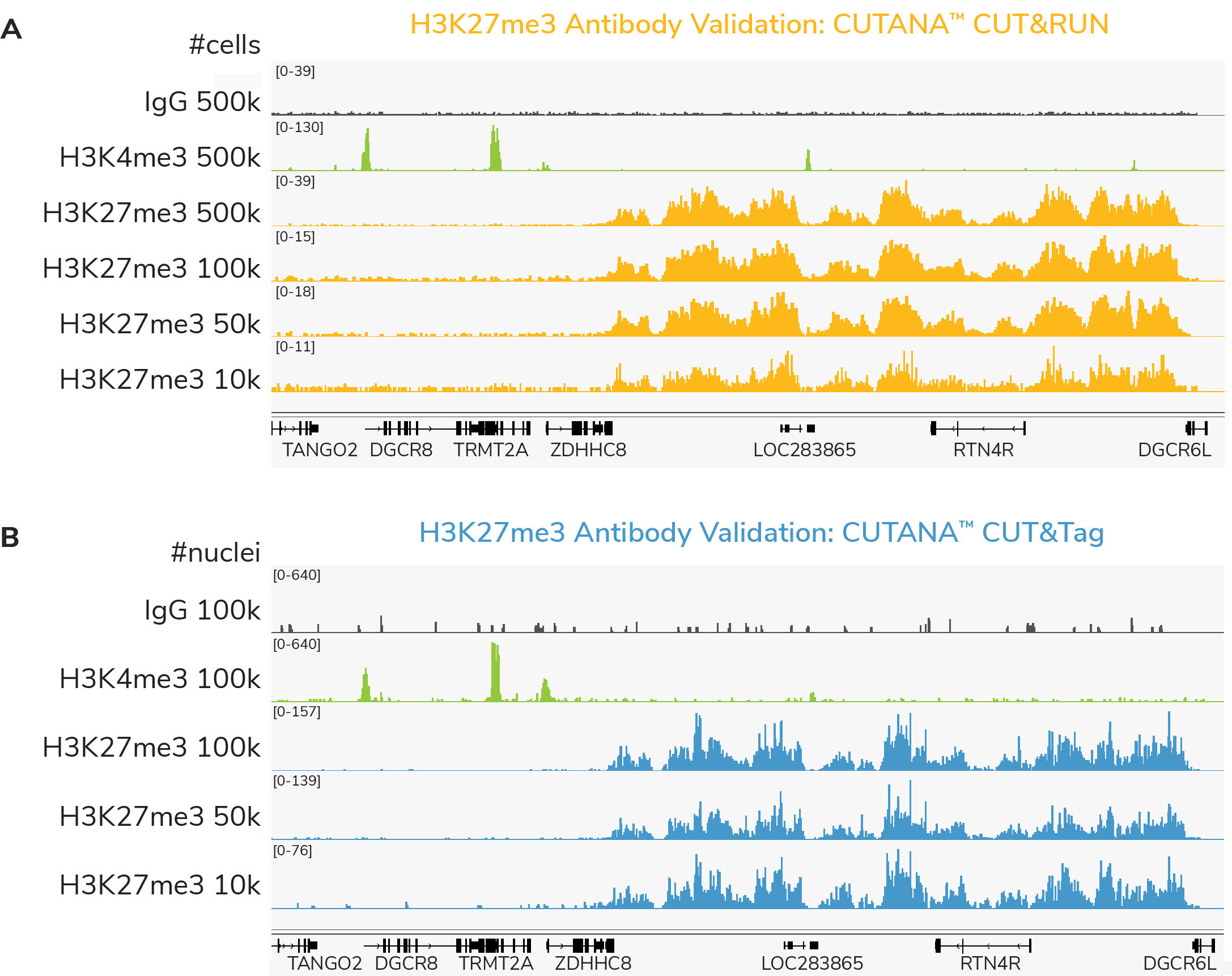
EpiCypher’s CUTANA™ CUT&RUN and CUT&Tag technologies are enabling major advancements in high-resolution profiling of histone post-translational modifications (PTMs) across scientific disciplines, including immunology, neuroscience, and cancer research1-3. Antibodies are crucial to the high sensitivity of CUT&RUN and CUT&Tag assays, where they are used to tether enzymes for target-specific cleavage and enrichment.
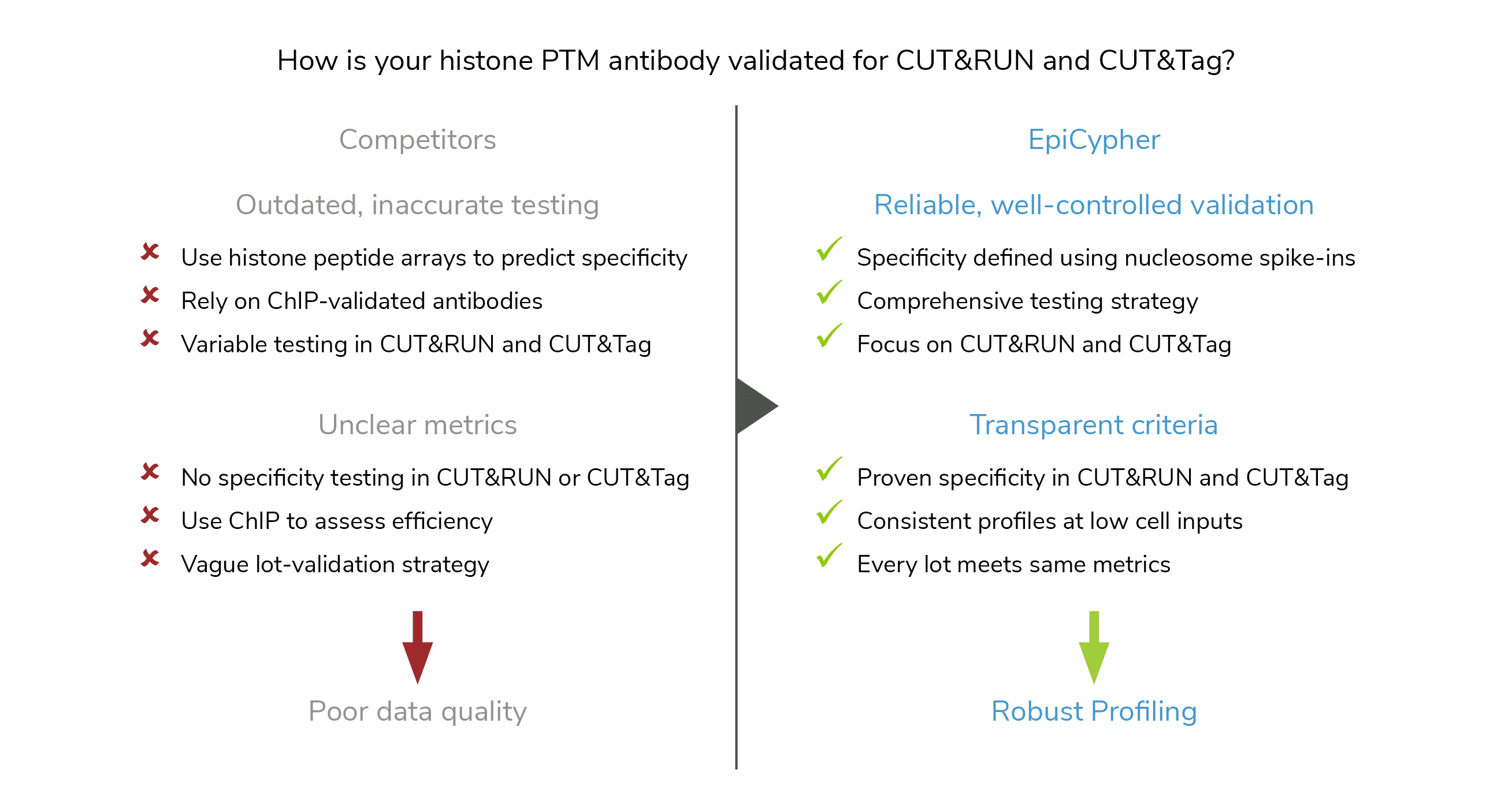
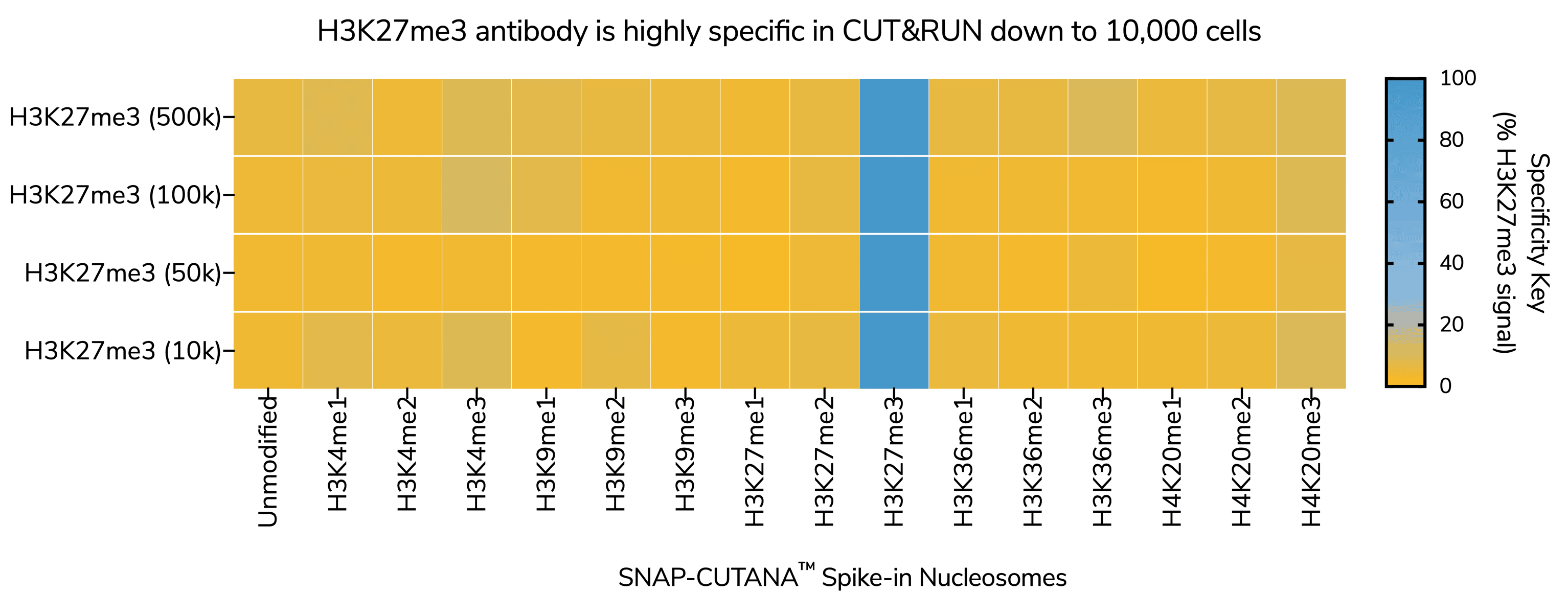
But not just any antibody will do! In fact, histone PTM antibodies are notoriously unreliable, with high rates of cross-reactivity and poor binding efficiency in chromatin mapping assays. Furthermore, EpiCypher found that histone PTM antibody performance can vary widely between assay types and substrates (i.e. peptides vs. nucleosomes) – making assay-specific testing a must for the epigenetics field4.
To realize the full the potential of CUTANA assays for epigenetics research, EpiCypher developed SNAP-Certified™ antibodies for CUT&RUN and CUT&Tag. Here, we will explore why EpiCypher’s antibody validation pipeline results in better antibodies for histone PTM profiling compared to traditional strategies and address common questions to help you select the best antibody for your CUT&RUN and CUT&Tag experiment.