Part 3: Modeling Membrane Fusion with SNARE Protein Analogs
Click the links for Part 1 and Part 2 of this three-part series.
Membrane fusion is a universal process in living organisms and is critical to proper cellular function. Examples of membrane fusion events include exocytosis, fertilization, envelopment of infecting viruses, and transport of proteins through the Golgi stack. Though essential, membrane fusion is not spontaneous because free energy is required to overcome steric hindrances, electrostatic repulsions, and hydration shells between two membranes as they move toward each other prior to fusion.
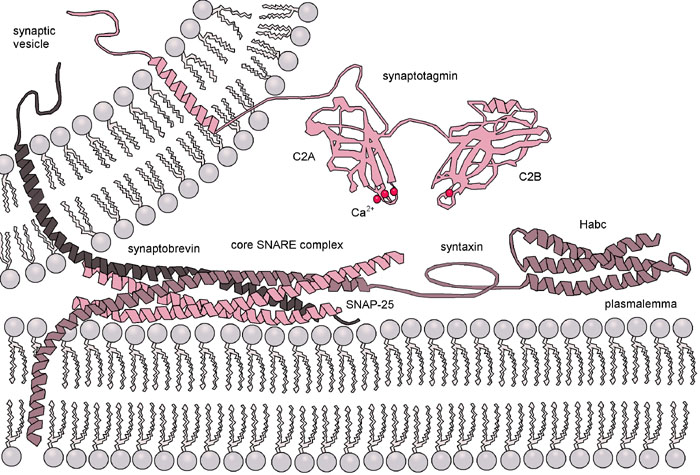
In cells membrane fusion is facilitated by a protein superfamily known as SNARE (for Soluble N-ethylmaleimide-sensitive-factor Attachment Receptor), of which there are 36 members in humans. The best studied SNARE-mediated membrane fusion process is exocytosis in neural cells. Indeed, the first proteins of the human SNARE family to be identified were synaptobrevin, syntaxin, and synaptosome-associated protein 25 (SNAP-25). Figure 1 shows a SNARE-mediated exocytosis.