Click Chemistry[1] describes pairs of functional groups that rapidly and selectively react (“click”) with each other under mild, aqueous conditions. The concept of Click Chemistry has been transformed into convenient, versatile and reliable two-step coupling procedures of two molecules A and B[1-5] that are widely used in biosciences[6-8], drug discovery[9] and material science[10].
Click Reagents by Application
... in Posttranslational Modification Analysis
Advantages of Click Chemistry:
- Highly selective, low background labeling:
CLICK-functional groups are inert to naturally occurring functional groups (“bioorthogonal”) such as amines - Rapid and quantitative labeling
- Allows non-radioactive analysis of enzymatic activities both in vitro and in vivo:
Small-sized CLICK functional groups possess excellent substrate properties
Click Reagents for Application on DNA
Cell proliferation is characterized by de novo DNA synthesis during the S-Phase of the cell cycle. A common approach for DNA synthesis monitoring relies on incorporation of ethynyl-labeled deoxyuridine (5-EdU[1,2,3,4,6]) or ethynyl-labeled deoxycytidine (5-EdC[5]) instead of their natural counterpart thymidine. This traditional method however, is hampered by cytotoxicity of Cu(I) which is generated during copper-catalyzed labeling of 5-EdU/5-EdC with Azide-functionalized detection molecules (fluorescent Azides, Azides of (Desthio)Biotin or FLAG Azide) (Fig. 1A).
Rieder et al.[7] reported now a copper-free method for DNA synthesis monitoring using incorporation of 5-Vinyl-2′-deoxyuridine (5-VdU) (instead of 5-EdU) into the DNA of proliferating cells (Fig. 1B). Labeling of the resulting Vinyl-functionalized DNA is achieved via Copper-free Alkene-Tetrazine Ligation that allows to introduce a Biotin group for subsequent purification (via Tetrazine-containing Biotinylation Reagents) or a fluorescent group for subsequent microscopic imaging (via Tetrazine-containing Fluorescent Dyes).
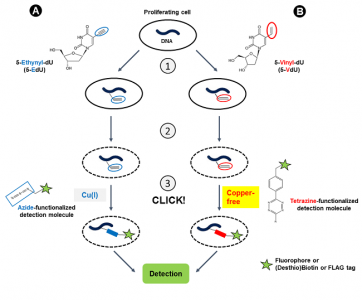
Figure 1: DNA synthesis monitoring with 5-EdU (A) and 5-VdU (B) is performed in a 3-step procedure:
1) Intracellular Ethynyl- or Vinyl-functionalization of DNA by incubation with 5-EdU or 5-VdU, respectively.
2) Cell fixation & permeabilization,
3) CLICK labeling with an Azide-functionalized detection molecule via CuAAC reaction (A)(the labeling procedure with 5-EdC is similar) or with a Tetrazine-functionalized detection molecule via copper-free Alkene-Tetrazine ligation (B). CuAAC: Copper (I) (Cu(I))-catalyzed terminal Alkyne-Azide Click Chemistry reaction.
| Products & Ordering | |
| 5-Ethynyl-2′-deoxyuridine (5-EdU) CLK-N001 | 7-Deaza-7-ethynyl-2′-deoxyadenosine (EdA) CLK-099 |
| 5-Vinyl-2′-deoxyuridine (5-VdU) CLK-050 5-Ethenyl-2′-deoxyuridine | CuAAC Cell Reaction Buffer Kit (THPTA based) CLK-074 |
| 5-Dibenzylcyclooctyne-2′-deoxyuridine (5-DBCO-dU) CLK-082 5-Dibenzylcyclooctyne-dU | CuAAC Cell Reaction Buffer Kit (BTTAA based) CLK-073 |
| 5-Azidomethyl-2′-deoxyuridine (5-AmdU) CLK-064 | CuAAC Reaction Ligand Test Kit (THPTA & BTTAA based) CLK-075 |
| 5-Ethynyl-2′-deoxycytidine (5-EdC) CLK-N003 | (2’S)-2′-Deoxy-2′-fluoro-5-ethynyluridine (F-ara-EdU) CLK-1403 |
Selected References
[1] Salic et al. (2008) A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. USA 105:2415.
[2] Li et al. (2010) Fluorogenic “click” reaction for labeling and detection of DNA in proliferating cells. Biotechniques 49 (1):525.
[3] Diermeier-Daucher et al. (2009) Cell Type Specific Applicability of 5-Ethynyl-2′-deoxyuridine (EdU) for Dynamic Proliferation Assesment in Flow Cytometry.Cytometry A 75A:535.
[4] Darzynkiewicz et al. (2011) Cytometry of DNA Replication and RNA synthesis: Historical Perspective and Recent Advances Based on “Click Chemistry”. Cytometry A 79A:328.
[5] Guan et al. (2011) Intracellular detection of cytosine incorporation in genomic DNA by using 5-ethynyl-2′-deoxycytidine. ChemBioChem 2 (14):2184.
[6] Diermeier-Daucher et al. (2010) Dynamic Proliferation Assessment in Flow Cytometry. Curr. Protoc. Cell Biol. 48:8.6.1.
[7] Rieder et al. (2014) Alkene-Tetrazine Ligation for Imaging Cellular DNA. Angew. Chem. Int. Ed. 53:9168.
Table 1: Enzymatic incorporation of CLICK-functionalized nucleotides.
PCR: Polymerase chain reaction with Taq Polymerase (CLK-T09, NU-1705) or family B polymerases Pwo, Deep Vent exo– and KOD XL (CLK-T05/-T06/-T07); NT: Nick Translation with DNAse I / DNA Polymerase I;
Primer Extension: Primer Extension with family B polymerases Pwo, Deep Vent exo– and KOD XL; 3′-End Labeling: Incorporation with Terminal deoxynucleotidyl Transferase (TdT); RT:: Reverse Transcription with Moloney Murine Leukemia Virus Reverse Transcriptase (MMLV RT)
| DNA Labeling | cDNA Labeling |
|||||
| Nucleotide | Cat. No. | PCR | NT | Primer Extension | 3’ End Labeling | RT |
| dUTP | ||||||
| 5-DBCO-PEG4-dUTP | CLK-048 |  1 1 |
n/a | n/a | n/a | n/a |
| 5-Azidomethyl-dUTP | CLK-084 | n/a | n/a | n/a | n/a | n/a |
| Azide-PEG4-Aminoallyl-dUTP | NU-1705 |  1 1 |
n/a | n/a |  2 2 |
n/a |
| 5-Ethynyl-dUTP | CLK-T07 |  1,2 1,2 |
n/a |  2 2 |
n/a | n/a |
| C8-Alkyne-dUTP | CLK-T05 |  1,2 1,2 |
n/a |  2 2 |
n/a | n/a |
| 5-TCO-PEG4-dUTP | CLK-035 |  1 1 |
n/a | n/a | n/a | n/a |
| 5-Vinyl-dUTP | CLK-068 |  1 1 |
n/a | n/a | n/a | n/a |
| dCTP | ||||||
| C8-Alkyne-dCTP | CLK-T06 |  1,2 1,2 |
n/a |  2 2 |
 2 2 |
n/a |
| 5-DBCO-PEG4-dCTP | CLK-060 |  1 1 |
n/a | n/a | n/a | n/a |
| 5-Azido-PEG4-dCTP | CLK-070 |  1 1 |
n/a | n/a | n/a | n/a |
| dATP | ||||||
| N6-Azidohexyl-dATP | CLK-NU-002 |  1 1 |
n/a | n/a | n/a | n/a |
1: in-house tested
2: external reference (see datasheet)
n/a: no data available
– : not applicable
5-hydroxymethylcytosine (5-hmC) is a DNA modification that is – along with its analog 5-methylcytosine (5-mC) – an important epigenetic marker associated with critical cellular functions such as stem cell differentiation and brain development[1]. A specific detection of 5-hmC however, is not trivial since traditional sodium bisulfite sequencing does not distinguish between 5-mC and 5-hmC sites[2].
Song et al.[3] reported an approach for the selective detection of 5-hmC residues in genomic DNA based on UDP-6-azide-glucose (UDP-6-N3-Glc) (Fig. 1). T4 Phage β-glucosyltransferase selectively transfers an azide-modified glucose moiety to the hydroxyl group of 5-hmC allowing detection via Copper-free strain-promoted alkyne-azide click chemistry (SPAAC) by
- introduction of a (Desthio)Biotin group (DBCO-containing (Desthio)Biotin) for subsequent purification tasks [1,2] or
- introduction of a fluorescent group (DBCO-containing Fluorescent Dyes ) for subsequent microscopic imaging [1-3].
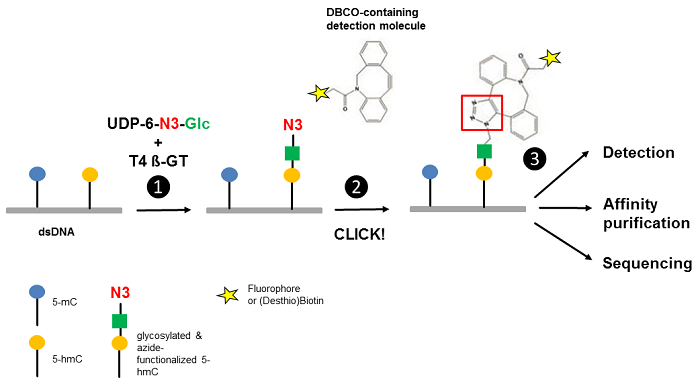 Figure 1 Workflow of 5-hmC detection. Step 1: T4 Phage ß-glycosyltransferase (T4 ß-GT)-mediated glycosylation and simultaneous azide-functionalization of 5-hmC residues using UDP-6-azide-glucose (UDP-6-N3-Glc). Step 2: Copper-free labeling of Azide (N3) with DBCO-containing detection molecule (DBCO-containing (Desthio)Biotin or DBCO-containing Fluorescent Dyes ) forming a stable triazole moiety. Step 3: Detection, affinity purification or sequencing of 5-hmC containing DNA (modified according to [3]).
Figure 1 Workflow of 5-hmC detection. Step 1: T4 Phage ß-glycosyltransferase (T4 ß-GT)-mediated glycosylation and simultaneous azide-functionalization of 5-hmC residues using UDP-6-azide-glucose (UDP-6-N3-Glc). Step 2: Copper-free labeling of Azide (N3) with DBCO-containing detection molecule (DBCO-containing (Desthio)Biotin or DBCO-containing Fluorescent Dyes ) forming a stable triazole moiety. Step 3: Detection, affinity purification or sequencing of 5-hmC containing DNA (modified according to [3]).
| Products & Ordering |
| UDP-6-azide-glucose CLK-076 |
Selected References
[1] Branco et al. (2012) Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nature Reviews Genetics 13:7.
[2] Huang et al. (2010) The behaviour of 5-hydroxymethycytosine in bisulfite sequencing. PLOS One 5(1):e8888.
[3] Song et al. (2011) Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nature Biotech 29(1):68.
[4] Li et al. (2012) Selective Capture of 5-hydroxymethylcytosine from Genomic DNA. J. Vis. Exp. 68:e4441.
[5] Song et al. (2016) Simultaneous single-molecule epigenetic imaging of DNA methylation and hydroxymethylation. PNAS 113(16):4339.
Click Reagents for Application on RNA
Labeling of RNA through bioorthogonal reactions is typically split into two main-steps: The metabolic incorporation (in vivo) or enzymatic installation (in vitro) of the reporter group into RNA and the chemical reaction with tagged probes for detection.
A common approach for RNA synthesis monitoring relies on incorporation of ethynyl-labeled uridine and subsequent labelling with Azide-functionalized detection molecules (fluorescent Azides, Azides of (Desthio)Biotin or FLAG Azide)(Fig. 1A)[1,2]. This Copper-catalyzed Azide-Alkyne Cycloaddition (CuAAC) is widely applied due to its fast kinetics and high efficiency, but is hampered by cytotoxicity of in situ generated Cu(I). The amount of Cu(I) can be reduced by using stabilizing ligands.
An attractive copper-free alternative to measure de novo RNA synthesis in proliferating cells is presented by using vinyl-modified nucleosides as chemical reporter, such as 5-Vinyl-uridine (5-VU)[3]. Labeling of the resulting vinyl-functionalized RNA is achieved via Inverse-Electron Demand Diels-Alder reaction (IEDDA) that allows for subsequent introduction of either a Biotin group for purification (via Tetrazine-containing Biotinylation Reagents) or a fluorescent group for subsequent microscopic imaging (via Tetrazine-containing Fluorescent Dyes).
Recently, the marked efficiency of Alkene-Tetrazine Ligation was demonstrated by pairing the properties of 2-Vinyladenosine (2-VA) or 7-Deaza-vinyl-adenosine (7-dVA), respectively with highly reactive Pyrimidyl tetrazines (Fig. 1B)[4].
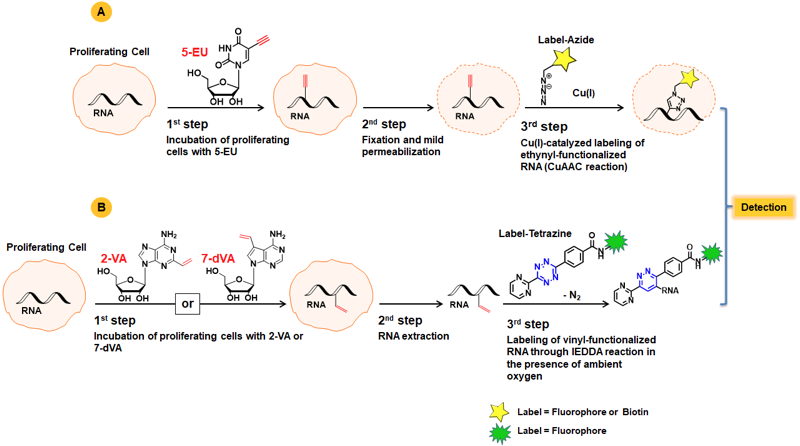
Figure 1: A) Workflow of RNA synthesis monitoring with 5-EU using copper-catalyzed Alkyne-Azide Ligation. B) Schematic overview of RNA labeling using vinyl-functionalized nucleosides and imaging through bioorthogonal IEDDA reaction with Pyrimidyl tetrazines.
| Products & Ordering | ||
| 5-Azidomethyl-uridine (5-AmU) CLK-063 | 5-Ethynyl-cytidine (5-EC) CLK-087 | CuAAC Cell Reaction Buffer Kit (THPTA based) CLK-074 |
| 5-Ethynyl-uridine (5-EU) CLK-N002 | 2-Vinyl-adenosine (2-VA) CLK-106 | CuAAC Cell Reaction Buffer Kit (BTTAA based) CLK-073 |
| 5-Vinyl-uridine (5-VU) CLK-049 5-Ethenyl-uridine | 7-Deaza-vinyl-adenosine (7-dVA) CLK-107 | CuAAC Reaction Ligand Test Kit (THPTA & BTTAA based) CLK-075 |
Selected References
[1] Jao et al. (2008) Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. USA 105 (41):15779.
[2] Darzynkiewicz et al. (2011) Cytometry of DNA Replication and RNA Synthesis: Historical Perspective and Recent Advances Based on “Click Chemistry”. Cytometry A 79A:328.
[3] Liu et al. (2019) A Nucleoside Derivative 5‑Vinyluridine (VrU) for Imaging RNA in Cells and Animals. Bioconjugate Chem. 30:2958.
[4] Kubota et al. (2019) Expanding the Scope of RNA Metabolic Labeling with Vinyl Nucleosides and Inverse Electron-Demand Diels−Alder Chemistry. ACS Chem. Biol. 14:1698.
References
[1] Hida et al. (2017) EC-tagging allows cell type-specific RNA analysis. Nucleic Acids Res. 45 (15):e138.
CLICK-functionalized adenosine (triphosphate): Analysis of newly synthesized poly(A) tails in vivo and in vitro
Poly(A) tails, a long chain of adenosine nucleotides of variable length, are posttranscriptionally added to the 3′-end of eukaryotic mRNAs by poly(A) polymerase (polyadenylation). This modification plays a critical role in mRNA metabolism and thus gene regulation e.g. by influencing mRNA stability, nuclear export and translation efficiency[1]. Traditional methods for the analysis of poly(A) tails do not allow the discrimination between preexisting and newly polyadenylated mRNA transcripts e.g. induced by changes in signaling pathways.
Two CLICK-functionalized adenosine analogs – N6-Propargyl-Adenosine (N6pA)[2] and 2-Ethynyl-Adenosine (2-EA)[3] have been reported as suitable substrates for poly(A) tail synthesis (polyadenylation) monitoring in mammalian cells. N6pA and 2-EA are cell permeable and incorporate instead of their natural analog adenosine both transcriptionally by RNA polymerase I, II and III and post-transcriptionally by poly(A) polymerase (Figure 1A). In vitro polyadenylation can be performed using the ATP analogs N6-Propargyl-ATP (N6pATP) or 2-Ethynyl-ATP (2-EATP) that serve as substrates for poly(A) polymerase from cell extracts[2] (Figure 1B).
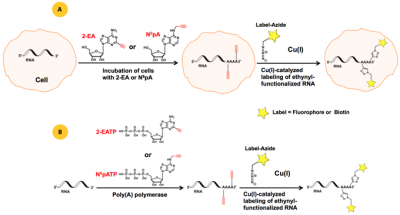
Figure 1: A) Principle of in vivo polyadenylation with the cell-permeable ethynyl-functionalized analogs 2-EA or N6pA[3], respectively. B) Principle of in vitro polyadenylation with ethynyl-functionalized ATP analogs 2-EATP[2] or N6pATP[3], respectively. The resulting ethynyl-functionalized RNA (population) can subsequently be labeled via Cu(I)-catalyzed click reactions, allowing introduction of a Biotin group for subsequent purification tasks (via Azides of Biotin) or a fluorescent group for subsequent microscopic imaging (via fluorescent Azides).
Selected References
[1] Colgan et al. (1997) Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11:2755.
[2] Grammel et al. (2012) Chemical Reporters for Monitoring RNA synthesis and Poly(A) Tail dynamics. ChemBioChem 13:1112.
[3] Curanovic et al. (2013) Global profiling of stimulus-induced polyadenyation in cells using a poly(A) trap. Nature Chemical Biology 9:671.
| Products & Ordering | |
| N6-Propargyl-adenosine CLK-N004 | CuAAC Cell Reaction Buffer Kit (THPTA based) CLK-074 |
| N6-Propargyl-ATP (N6pATP) CLK-NU-001 | CuAAC Cell Reaction Buffer Kit (BTTAA based) CLK-073 |
| 2-Ethynyl-adenosine CLK-N005 | CuAAC Reaction Ligand Test Kit (THPTA & BTTAA based) CLK-075 |
| 2-Ethynyl-ATP (2-EATP) CLK-NU-004 | CuAAC Biomolecule Reaction Buffer Kit (BTTAA based) CLK-071 |
| 2-Vinyl-adenosine (2-VA) CLK-106 | CuAAC Biomolecule Reaction Buffer Kit (THPTA based) CLK-072 |
| 7-Deaza-vinyl-adenosine (7-dVA) CLK-107 |
Table 1: Enzymatic incorporation of CLICK-functionalized nucleotides.
| cRNA Labeling by in vitro Transcription with… |
3′-RNA Labeling by… | ||||||
| Nucleotide | Cat. No. | T7 RNA Polymerase | T3 RNA Polymerase | SP6 RNA Polymerase | yPAP | TdT | T4 RNA Ligase |
| UTP | |||||||
| 5-Azidomethyl-UTP | CLK-083 |  1 1 |
n/a | n/a | n/a | n/a | n/a |
| 5-Azido-C3-UTP | NU-157 |  1,2 1,2 |
n/a | n/a | – | – | – |
| 5-Azido-PEG4-UTP | CLK-054 |  1 1 |
n/a | n/a | n/a | n/a | n/a |
| 5-Ethynyl-UTP | CLK-T08 |  1 1 |
n/a | n/a | – | – | – |
| DBCO-PEG4-UTP | CLK-055 |  1 1 |
n/a | n/a | n/a | n/a | n/a |
| Vinyl-UTP | CLK-069 |  1 1 |
n/a | n/a | n/a | n/a | n/a |
| ATP | |||||||
| 8-Azido-ATP | NU-155 | – | – | – |  2 2 |
– | – |
| 3′-Azido-2’,3′-ddATP | NU-882 | – | – | – |  2 2 |
– | – |
| CTP | |||||||
| 5-Azido-PEG4-CTP | CLK-053 |  1 1 |
n/a | n/a | n/a | n/a | n/a |
| 5-DBCO-PEG4-CTP | CLK-056 |  1 1 |
n/a | n/a | n/a | n/a | n/a |
| 3′-dATP (Cordycepin derivative) | |||||||
| N6-Azidohexyl-3′-dATP | NU-1707 | – | – | – | n/a | – | – |
| Cytidine-5’/3′-phosphate | |||||||
| pCp-Azide | NU-1708 | – | – | – | – | – |  2 2 |
| CpG | |||||||
| 5-DBCO-PEG4-dCpG | CLK-062 | n/a | n/a | n/a | n/a | n/a | n/a |
yPAP: Yeast Poly A Polymerase
TdT: Terminal deoxynucleotidyl Transferase.
1: in-house tested
2: external reference (see datasheet)
n/a: no data available
– : not applicable
| Products & Ordering | |
| 5-Azidomethyl-UTP CLK-083 | 3′-Azido-2’,3′-ddATP NU-882 |
| 5-Azido-C3-UTP NU-157 | 5-Azido-PEG4-CTP CLK-053 |
| 5-Azido-PEG4-UTP CLK-054 | 5-DBCO-PEG4-CTP CLK-056 |
| 5-Ethynyl-UTP (5-EUTP) CLK-T08 | N6-(6-Azido)hexyl-3′-dATP NU-1707 (Cordycepin triphosphate derivative) |
| 5-DBCO-PEG4-UTP CLK-055 | pCp-Azide NU-1708 |
| 5-Vinyl-UTP CLK-069 | 5-DBCO-PEG4-dCpG CLK-062 |
| 8-Azido-ATP NU-155 | C8-Alkyne-dCTP CLK-T06 |
Click Reagents for Application on Proteins
O-Propargyl-puromycin: Non-radioactive monitoring of nascent protein synthesis
The dynamics of global protein synthesis (both spatial and temporal) is an essential parameter to characterize the cellular response under various physiological and pathological conditions. Current studies of the cellular protein level rely on indirect methods such as DNA and mRNA microarrays or classical radioactive metabolic labeling with 35S-methionine.
Liu et al. reported a non-radioactive alternative to analyze newly synthesized proteins in cell culture and whole organisms that is based on an alkyne analog of puromycin (Fig. 1). The cell-permeable O-propargyl-puromycin (Fig. 1A) incorporates into the C-terminus of translating polypeptide chains thereby stopping translation.
The resulting C-terminal alkyne labeled proteins can be detected via Cu(I)-catalyzed click chemistry that offers the choice to introduce a Biotin group (Azides of Biotin) for subsequent purification tasks or a fluorescent group (Azides of fluorescent dyes) for subsequent microscopic imaging[1,2,3].
In contrast to previously reported non-radioactive methionine analog-approaches[4,5], methionine free-medium is not required for O-propargyl-puromycin-based monitoring of nascent protein synthesis.
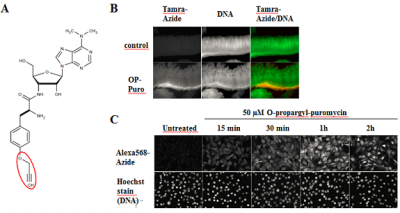
Figure 1: O-propargyl-puromycin labels newly synthesized proteins in cell culture and whole organisms (modified according to [1]. A) Chemical structure of O-propargyl-puromycin (OP-Puro). Visualization of incorporated OP-puro is performed via Cu(I)-catalyzed Click Chemistry (CuAAC) with Azides of fluorescent dyes or Azides of Biotin. B) Nascent protein expression in crypts of mouse small intestine was visualized by whole-mount staining. O-Propargyl-puromycin labeled proteins were detected with Tamra-azide (red) and nuclear DNA was stained with OliGreen (green) (modified according to [1]) C) Newly synthesized proteins in NIH3T3 cells are rapidly detected by incubation with 50 µM OP-puro. OP-puro labeled proteins have been visualized by AzDye568-azide, nuclear DNA was stained with Hoechst dye (modified according to [1]).
Selected References
[1] Liu et al. (2012) Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc. Natl. Acad. Sci. USA 109 (2):413.
[2] Goodman et al. (2012) Imaging of protein synthesis with puromycin. Proc. Natl. Acad. Sci. USA 109 (17):E989.
[3] Salic et al. (2012) Reply to Goodman et al.: Imaging protein synthesis with puromycin and the subcellular localization of puromycin-polypeptide conjugates. Proc. Natl. Acad. Sci. USA 109 (17):E990.
[4] Dieterich et al. (2010) In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nature Neuroscience 13 (7):897.
[5] Dieck et al. (2012) Metabolic Labeling with Noncanonical Amino Acids and Visualisation by Chemoselective Fluorescent Tagging. Current Protocols in Cell Biology 7:7.11.1.
The dynamics of global protein synthesis (both spatial and temporal) is an essential parameter to characterize the cellular response under various physiological and pathological conditions. Protein synthesis has traditionally been monitored by metabolic labeling with 35S-methionine that competes with natural methionine for the incorporation into newly synthesized proteins. The detection is limited to autoradiography.
The cell-permeable methionine-analogs 4-Azido-L-homoalanine (L-AHA) and L-Homopropargylglycine (HPG) provide a non-radioactive alternative to analyze the global protein synthesis in cell culture. They are randomly incorporated instead of methionine during translation leading to full-length, azide-labeled or alkyne-modified proteins, respectively[1,2,3](Fig. 1).
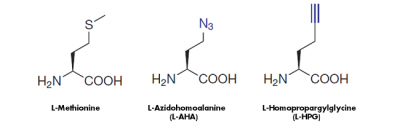
Figure 1: Methionine analogs are suitable to incorporate azide groups (L-AHA) or alkyne groups (L-HPG) into nascent proteins. Visualization of the incorporated azide groups (L-AHA-based labeling) is performed via Cu(I)-catalyzed Click Chemistry with Alkynes of fluorescent dyes or Alkynes of Biotin or via Cu-free Click Chemistry with DBCO-containing fluorescent dyes or DBCO-containing Biotin. Visualiation of Alkyne groups (HPG-based labeling) can be performed with Cu(I)-catalayzed Click Chemistry only.
| Products & Ordering | |
| 4-Azido-L-homoalanine HCl (L-AHA) CLK-AA005 (S)-2-Amino-4-azidobutanoic acid hydrochloride | CuAAC Cell Reaction Buffer Kit (BTTAA based) CLK-073 |
| L-Homopropargylglycine (L-HPG) CLK-1067 | CuAAC Reaction Ligand Test Kit (THPTA & BTTAA based) CLK-075 |
| CuAAC Cell Reaction Buffer Kit (THPTA based) CLK-074 | L-Homopropargylglycine (L-HPG) CLK-1067 |
Selected References
[1] Dieck et al. (2012) Metabolic Labeling with Noncanonical Amino Acids and Visualisation by Chemoselective Fluorescent Tagging. Current Protocols in Cell Biology 7:7.11.1.
[2] Kiick et al. (2002) Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc. Natl. Acad. Sci. USA 99 (1):19.
[3] Dieterich et al. (2010) In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nature Neuroscience 13 (7):897.
CLICK-functionalized NHS Ester or Maleimides enable the introduction of an azide-, alkyne-, DBCO-, tetrazine- or trans-cyclooctene-group into recombinant proteins via targeting of lysine groups (NHS Ester-based Amine Labeling) or cysteine groups (Maleimide-based Thiol Labeling) (Tab. 1). The resulting randomly CLICK-functionalized protein can subsequently be processed via Cu(I)-catalyzed (azide-alkyne) or Cu(I)-free (azide-DBCO /tetrazine/TCO) click chemistry that offers the choice
- to introduce a Biotin group for subsequent purification tasks (via Azides of Biotin, Alkynes of Biotin or DBCO-containing Biotinylation reagents)
- to introduce a fluorescent group for subsequent microscopic imaging (via fluorescent Azides, fluorescent Alkynes, DBCO-containing fluorescent dyes or Tetrazine-containing fluorescent dyes)
- to crosslink the protein to complementary CLICK-functionalized biomolecules e.g. DNA
Table 1: Overview of CLICK-functionalized NHS ester and Maleimides for Amino & Thiol Labeling, respectively.
| Reactive groups | NHS Ester | Maleimide |
| Azide | Azide-PEG4-NHS ester | Azide-PEG3-Maleimide |
| Alkyne | Propargyl-NHS ester Acetylene-PEG4-NHS ester | Propargyl-Maleimide Acetylene-PEG4-Maleimide |
| DBCO | DBCO-NHS ester DBCO-PEG4-NHS ester DBCO-PEG-Sulfo-NHS ester | DBCO-Maleimide DBCO-PEG4-Maleimide |
| Tetrazine | 6-Methyl-Tetrazine-PEG5-NHS ester | 6-Methyl-Tetrazine-PEG4-Maleimide |
| Trans-Cyclooctene (TCO) | TCO-NHS ester TCO-PEG4-NHS ester |
| Products & Ordering | |
| Azido-PEG4-NHS ester CLK-AZ103 | DBCO-PEG4-NHS ester CLK-A134 Dibenzylcyclooctyne-PEG4-NHS ester |
| Azido-PEG3-Maleimide Preparation Kit CLK-AZ107 | DBCO-Sulfo-NHS ester (sodium salt) CLK-A124 Sulfo-Dibenzylcyclooctyne-NHS ester |
| Propargyl-NHS ester CLK-TA111 | DBCO-Maleimide CLK-A108 Dibenzylcyclooctyne-Maleimide |
| Acetylene-PEG4-NHS ester CLK-TA103N | DBCO-PEG4-Maleimide CLK-A108P Dibenzylcyclooctyne-PEG4-Maleimide |
| Propargyl-Maleimide CLK-TA113 | 6-Methyl-Tetrazine-PEG5-NHS ester CLK-A138 |
| Acetylene-PEG4-Maleimide CLK-TA104 | TCO-NHS ester CLK-1016 trans-Cyclooctene-NHS ester |
| DBCO-NHS ester CLK-A133 Dibenzylcyclooctyne-NHS ester | TCO-PEG4-NHS ester CLK-A137 trans-Cyclooctene-PEG4-NHS ester |
Azide-, Alkyne-, DBCO-, TCO- and Tetrazine-functionalized proteins are efficiently immobilized on complementary functionalized matrizes (agarose or magnetic beads) via click chemistry based reaction procedures.
A tool-box of CLICK-functionalized matrizes is available to fit your specific application & molecule requirements (Table 1).
Table 1: Overview of available CLICK-functionalized agaroses & magnetic beads.
| Purification of | Agarose | Magnetic Beads |
| Alkyne- or DBCO-functionalized proteins | Azide Agarose Click Chemistry Capture Kit | Azide Magnetic Beads |
| Azide-functionalized proteins | Alkyne Agarose DBCO Agarose Click Chemistry Capture Kit | Alkyne Magnetic Beads DBCO Magnetic Beads |
| TCO-functionalized proteins | Methyl-Tetrazine Agarose | |
| Tetrazine-functionalized proteins | TCO Agarose |
| Products & Ordering | |
| Azide Agarose CLK-1038 | PC Alkyne Agarose CLK-1142 |
| TCO Agarose CLK-1198 | Dde Alkyne Agarose CLK-1140 |
| Methyl-Tetrazine Agarose CLK-1199N | DBCO Agarose CLK-1034 |
| Azide Magnetic Beads CLK-1036 | DBCO Magnetic Beads CLK-1037 |
| Alkyne Agarose CLK-1032 | Click Chemistry Capture Kit CLK-1065 |
| Alkyne Magnetic Beads CLK-1035 |
Click Reagents for Application on Lipids
Choline-containing phospholipids e.g. phosphatidylcholine or sphingomyelin are a class of lipids that are major components of cell membranes. They are critical for structural membrane integrity & signaling as well as celluar metabolism. Hence, the dynamics of choline-containing phospholipid synthesis is an essential parameter to analyze the cellular response under both physiological and pathological conditions.
Jao et al.[1,2] and Caishun et al.[3] reported a Click Chemistry-based approach to label newly synthesized choline-containing phospholipids in cell culture and whole organisms: The choline analogs Propargyl-choline (PCho) and 1-Azidoethyl-choline (AECho) are cell-permeable and metabolically incorporated into all classes of choline-containing phospholipids via the CDP-choline pathway.
| Products & Ordering | |
| Azide Agarose CLK-1038 | PC Alkyne Agarose CLK-1142 |
| TCO Agarose CLK-1198 | Dde Alkyne Agarose CLK-1140 |
| Methyl-Tetrazine Agarose CLK-1199N | DBCO Agarose CLK-1034 |
| Azide Magnetic Beads CLK-1036 | DBCO Magnetic Beads CLK-1037 |
| Alkyne Agarose CLK-1032 | Click Chemistry Capture Kit CLK-1065 |
| Alkyne Magnetic Beads CLK-1035 |
Selected References
[1] Jao et al. (2009) Metabolic Labeling and direct imaging of choline phospholipids in vivo. PNAS 106 (36):15332.
[2] Jao et al. (2015) Biosynthetic Labeling and Two-Color Imaging of Phospholipids in Cells. Chem. Bio. Chem. 16:472.
[3] Caishun et al. (2014) Practical Labeling Methodology for Choline-derived Lipids and Applications in Live Cell Fluorescence Imaging. Photochemistry and Photobiology 90:686.
Click Reagents for Posttranslational Modification Analysis
γ-phosphate CLICK-functionalized ATP analogs: Kinase substrate identification by non-radioactive in vitro Phosphorylation
Kinase substrates are routinely identified by in vitro phosphorylation with radioactive (“hot”) ATP whose modified gamma-phosphate (32 or 33γP) is transferred by a kinase to a serine, threonine or tyrosine hydroxyl group of its peptide/protein substrate.
Lee et al.[1] reported a non-radioactive version of in vitro phosphorylation were azide- and alkyne-γP-modified ATP analogs have been successfully used instead of 32γP-labeled ATP to phosphorylate p27kip1 with protein kinase cdk2 (Fig. 1). The phosphorylated substrate can subsequently be labeled with Biotin or fluorescent dyes via Cu(I)- catalyzed or Cu(I)-free Click-Chemistry (Tab. 1).
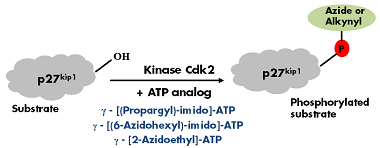
Figure 1: GST-tagged recombinant p27kip1 was incubated with kinase Cdk2 in the presence of one of the following Azide/Alkyne γP-labeled ATP analogs: γ-[(Propargyl)-imido]-ATP, γ-[(6-Azidohexyl)-imido]-ATP or γ-[2-Azidoethyl]-ATP. Phosphorylation was detected by immunoblotting with a phospho-p27kip1 specific antibody[1].
Table 1: Alkyne and Azide groups are detected via Cu(I)-catalyzed or Cu(I)-free Click Chemistry, respectively.
| ATP analog | Click Chemistry Type | Labeling with |
| γ-[(Propargyl)-imido]-ATP | Cu(I)-catalyzed | Azides of fluorescent dyes Azides of Biotin |
| γ-[(6-Azidohexyl)-imido]-ATP | Cu(I)-free | DBCO-containing fluorescent dyes DBCO-containing Biotin |
| γ-[2-Azidoethyl]-ATP |
| Products & Ordering | |
| γ-[(Propargyl)-imido]-ATP CLK-T11 | CuAAC Biomolecule Reaction Buffer Kit (BTTAA based) CLK-071 |
| γ-[(6-Azidohexyl)-imido]-ATP CLK-T12 | CuAAC Biomolecule Reaction Buffer Kit (THPTA based) CLK-072 |
| γ-(2-Azidoethyl)-ATP NU-1701 | CuAAC Reaction Ligand Test Kit (THPTA & BTTAA based) CLK-075 |
Selected References
[1] Lee et al. (2009) Synthesis and reactivity of novel y-phosphate modified ATP analogues. Bioorg Med Chem Lett. 19:3804.
N6 CLICK-functionalized ATP: AMP transferase substrate identification by non-radioactive in vitro AMPylation
AMPylation is a recently discovered post-translational modification that is characterized by the covalent modification of protein hydroxyl groups (serine, threonine or tyrosine) with adenylyl 5ʹ-monophosphate (AMP) catalyzed by specific AMP transferases[1,2]. The traditional method for the selective tagging of AMPylated proteins relies on a combination of radioactive labeling with 32P-alpha-ATP and targeted mass spectrometry.
Grammel et al.[1] reported a non-radioactive version of in vitro AMPylation were an Alkyne-modified ATP (N6-Propargyl-ATP (N6pATP)) has been sucessfully used to ampylate both purified proteins as well as proteins in cellular extracts. The resulting ampylated substrates can subsequently be labeled via Cu(I)-catalyzes click chemistry that offers the choice
-
- to introduce a Biotin group for subsequent purification tasks (via Azides of Biotin)
- to introduce a fluorescent group for subsequent microscopic imaging (via fluorescent Azides)
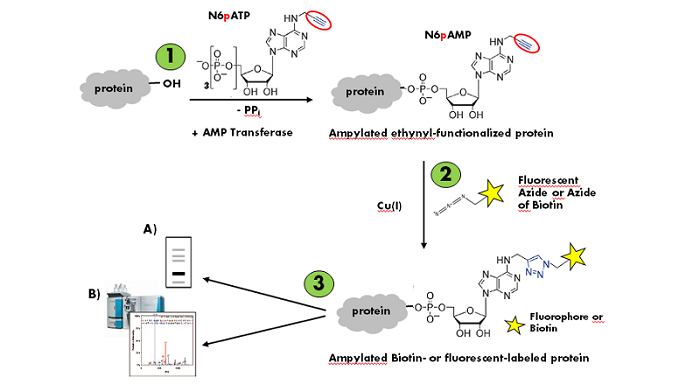
Figure 1: Workflow of in vitro AMPylation of proteins with N6-Propargyl-ATP (N6pATP) (modified according to [1]).
Step 1: Enzymatic AMPylation and simultaneous ethynyl-functionalization of the substrate protein. Step 2: Cu(I)-catalyzed Labeling of the ampylated ethynyl-functionalized substrate protein. Step 3: Detection via A) in-gel fluorescence analysis or B) mass spectrometry.
| Products & Ordering | |
| N6-Propargyl-ATP (N6pATP) CLK-NU-001 | CuAAC Biomolecule Reaction Buffer Kit (THPTA based) CLK-072 |
| CuAAC Biomolecule Reaction Buffer Kit (BTTAA based) CLK-071 | CuAAC Reaction Ligand Test Kit (THPTA & BTTAA based) CLK-075 |
Selected References
[1] Grammel et al. (2011) A Chemical Reporter for Protein AMPylation. J. Am. Chem. Soc. 133:17103.
[2] Broncel et al. (2012) A New Chemical Handle for Protein AMPylation at the Host-Pathogen Interface. ChemBioChem 13:183.
Glycosylation of proteins is a posttranslational modification that is characterized by the transfer of multiple monosaccharides (glycan structure) e.g. to serine/threonine residues (O-linked glycosylation), asparagine residues (N-linked glycosylation) or tryptophan residues (C-linked glycosylation).[1] The dynamics of global glycan synthesis is an essential parameter to characterize the cellular response under various physiological and pathological conditions[2] however, metabolic labeling studies of the cellular glycan level still primarily rely on classical radioactive labeling with 3H-monosaccharides.
CLICKable tetraacetylated derivatives of naturally occurring monosaccharides N-Acetylmannosamine (ManNAc), N-Acetylglucosamine (GlcNAc), N-Acetylgalactosamine (GalNAc) provide a non-radioactive alternative for metabolic glycan labeling[2-5] (Fig. 1).
Azide-functionalized monosaccharides (Ac4ManNAz, Ac4GlcNAz, Ac4GalNAz) facilitate the subsequent detection of the correspondingly functionalized proteins via Cu(I)-catalyzed Azide-terminal Alkyne or Cu(I)-free Azide-strained Alkyne click chemistry , respectively. Cyclopropene-functionalized monosaccharides (Ac4ManNCyoc, Ac4GlcNCyoc) are detected via Cyclopropene-Tetrazine reaction[6].
Azide- and Cyclopropene-functionalizd monosaccharides are ideally suited for dual-labeling approaches.[4,5]
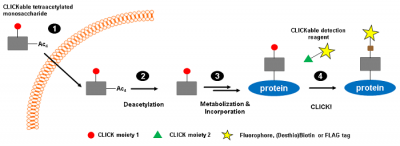
Figure 1: CLICKable monosaccharides are metabolically processed and subsequently incorporated instead of their natural counterparts into glycan structures of proteins (modified according to [3]).
Step 1: Intracelluar uptake of tetraacetylated CLICKable monosaccharide. Step 2: Deacetylation by endogenous esterases. Step 3: Metabolisation and incorporation into cellular glycan structures. Step 4: Detection of CLICK-functionalized proteins.
| Products & Ordering | |
| Ac4GlcNCyoc CLK-042 N-Cyclopropeneacetylatedglucosamine-tetraacylated | Ac4ManNCyoc CLK-041 N-Cyclopropeneacetylatedmannosamine-tetraacylated |
| Ac4GlcNAz CLK-1085 N-azidoacetylglucosamine-tetraacylated (Ac4GlcNAz) | Ac4ManNAz CLK-1084 N-azidoacetylatedmannosamine-tetraacylated (Ac4ManAz) |
| Ac4GalNAz CLK-1086 N-azidoacetylgalactosamine-tetraacylated (Ac4GalNAz) | CuAAC Reaction Ligand Test Kit (THPTA & BTTAA based) CLK-075 |
| CuAAC Cell Reaction Buffer Kit (BTTAA based) CLK-073 | CuAAC Cell Reaction Buffer Kit (THPTA based) CLK-074 |
Selected References
[1] Chauhan et al. (2013) In silico Platform for Prediction of N-, O- and C-Glycosites in Eukaryotic Protein Sequences. PLOS ONE 8(6):e67008.
[2] Laughlin et al. (2009) Imaging the glycome. Proc. Nat. Acad. Sci. 106 (1):12.
[3] Spaete et al. (2014) Expanding the scope of cyclopropene reporters for the detection of metabolically engineered glycoproteins by Diels-Alder reactions. Beilstein J. Org. Chem. 10:2235.
[4] Spaete et al. (2013) Rapid Labeling of Metabolically Engineered Cell-Surface Glycoconjugates with a Carbamate-Linked Cyclopropene Reporter. Bioconjugate Chem. 25:147.
[5] Patterson et al. (2014) Improved cyclopropene reporters for probing protein glycosylation. Mol. BioSyst. 10:1693.
[6] Patterson et al. (2012) Functionalized Cyclopropenes As Bioorthogonal Chemical Reporters. J. Am. Chem. Soc. 134:18638.
| Products & Ordering |
| 2-Ethynyl-Adenosine-NAD+ CLK-043 (2-EA-NAD+) |
Click Reagents for Application on Bacteria/Pathogens
Gram-negative bacteria are characterized by a lipopolysaccharide-containing outer cell membrane with 3-Deoxy-D-manno-octulosonic acid (KDO) as an essential component. The Azide-functionalized 3-Deoxy-D-manno-octulosonic acid (KDO-Azide) provides an attractive approach for the detection of gram-negative bacteria in living cultures[1]: It is cell-permeable, intracellularly processed and incorporated instead of its natural counterpart KDO.
Tested bacterial strains[1]:
Legionella pneumophila, Salmonella typhimurium, Escherichia coli O86, Escherichia coli K12
References
[1] Dumont et al. (2012) Click-Mediated Labeling of Bacterial Membranes through Metabolic Modification of the Lipopolysaccharide Inner Core. Angew. Chem. Int. Ed. 51:3143.
Legionaminic acid (LEG) is an essential lipopolysaccharide component exclusively synthesized by Legionella pneumophila. The Azide-functionalized Legionaminic acid precursor pLEG-Azide (Man2NAc4NAc6-N3) provides an attractive approach for the specific detection of Legionella pneumophila in living cultures: It is cell-permeable, intracellularly processed and incorporated instead of its natural counterpart.
Tested Legionella pneumophila strains[1]:
Legionella pneumophila SG1 (Paris), Legionella pneumophila SG1 (Lens), Legionella pneumophila SG1 (Philadelphia), Legionella pneumophila SG3, Legionella pneumophila SG4, Legionella pneumophila SG5, Legionella pneumophila SG6
References
[1] Mas Pons et al. (2014) Identification of Living Legionella pneumophila using species-specific metabolic lipopolysaccharide Labeling. Angew. Chem. Int. Ed. 53:1275.
Myobacteria are characterized by a trehalose-lipopolysaccharide-containing outer cell membrane. The Azide-functionalized 6-Azido-Trehalose provides an attractive approach for the detection of Myobacteria in living cultures[1]: It is cell-permeable, intracellularly processed and incorporated instead of its natural counterpart.
Myobacteria strains tested[1]:
Mycobacterium smegmatis mc2155, Mycobacterium tuberculosis H37Rv, Mycobacterium bovis BCG
References
[1] Swarts et al. (2012) Probing the Myobacterial Trehalome with Bioorthogonal Chemistry. J. Am. Chem. Soc. 134(39):16123.
Selected References
Introduction to the concept of Click Chemistry
[1] Kolb et al. (2001) Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. 40 (11):2004.
[2] Sletten et al. (2009) Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality. Angew. Chem. Int. Ed.48:6998.
[3] Jewett et al.(2010) Cu-free click cycloaddition reactions in chemical biology. Chem. Soc. Rev. 39 (4):1272.
[4] Best et al. (2009) Click Chemistry and Bioorthogonal Reactions: Unprecedented Selectivity in the Labeling of Biological Molecules. Biochemistry.48:6571.
[5] Lallana et al. (2011) Reliable and Efficient Procedures for the Conjugation of Biomolecules through Huisgen Azide–Alkyne Cycloadditions. Angew. Chem. Int. Ed. 50:8794.
Overview of Click Chemistry Applications
[6] Grammel et al. (2013) Chemical Reporters for biological discovery. Nature Chemical Biology 9:475.
[7] Xie et al. (2013) Cell-selective metabolic labeling of biomolecules with bioorthogonal functionalities. Current Opinion in Chemical Biology 17:747.
[8] Su et al. (2013) Target identification of biologically active small molecules via in situ methods. Current Opinion in Chemical Biology 17:768.
[9] Zeng et al. (2013) The Growing Impact of Bioorthogonal Click Chemistry on the Development of Radiopharmaceuticals. J Nucl Med 54:829.
[10] Evans et al. (2007) The Rise of Azide–Alkyne 1,3-Dipolar ‘Click’ Cycloaddition and its Application to Polymer Science and Surface Modification. Australian Journal of Chemistry 60 (6):384.
