Nucleotides for Application in Cell Cycle & Proliferation
Cell proliferation is characterized by de novo DNA synthesis during the S-Phase of the cell cycle as well as increased transcriptional activity accompanied by de novo RNA synthesis.
A common approach for monitoring global DNA and RNA synthesis relies on the incorporation of 5-labeled (deoxy) uridine analogs instead of their structural analogs (RNA: uracil; DNA: thymidine) into nascent DNA or RNA strands by cellular DNA and RNA polymerases, respectively. Non-phosphorylated labeled (deoxy)uridines (5-X-(d)U) are cell permeable and thus suitable for in vivo labeling (living cells)[1-5]. On the contrary, labeled (deoxy)nucleotide triphosphates (5-X-(d)UTP) are not cell permeable and can be used for in vitro DNA/RNA synthesis analysis (cell free extracts)[6,7] or needs to be delivered into cells for in vivo studies e.g. by lipofection or microinjection[8].
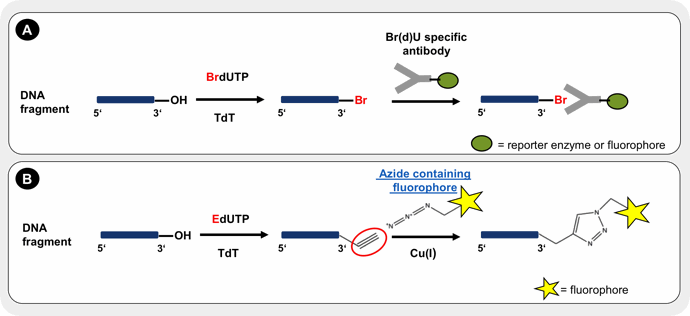
Traditional approaches rely on the incorporation of Bromine-labeled (deoxy)uridine (5-Br(d)U) or (deoxy)uridine triphosphates (5-Br(d)UTP) that are subsequently detected by BrdU specific antibodies (Tab. 1). There are however harsh permeabilization procedures required to achieve a sufficient staining efficiency which is due to the high molecular weight of the antibody (around 150.000 kDa) that limits the rate of antibody diffusion into the cell.
The incorporation of ethynyl-labeled (deoxy)uridine (5-E(d)U) provides a novel alternative for in vivo monitoring of DNA and RNA synthesis. The detection of 5-E(d)U is achieved by covalent conjugation of an azide-containing fluorophore → Click Chemistry via Cu(I) catalyzed alkyne-azide click chemistry (CuAAC) (Tab. 1) that are readily incorporated into cells under mild permeabilization conditions due their significantly lower size (<1000 kDa).
Table 1: Nucleotide selection guide for monitoring of global DNA and RNA synthesis.
| Measurement of de novo… | Cell permeable nucleosides | Non cell permeable nucleotides | Detection |
| …DNA synthesis | 5-BrdUTP[2] | BrdU specific antibody | |
| 5-EdU[3,4] | Azides of fluorescent dyes → Click Chemistry | ||
| …RNA synthesis | 5-BrUTP[6,7] | BrdU specific antibody | |
| 5-EU[8] | Azides of fluorescent dyes → Click Chemistry |