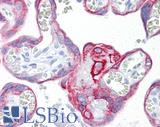
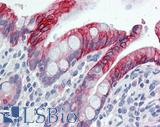
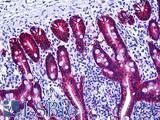
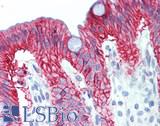
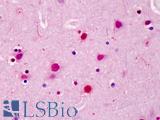
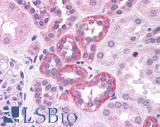
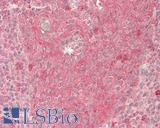
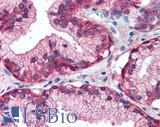
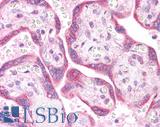
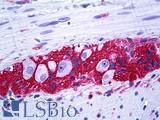
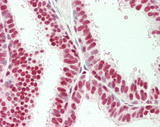
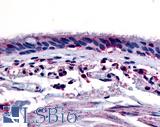
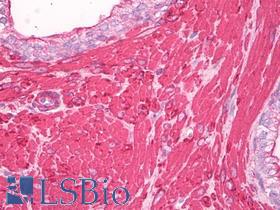
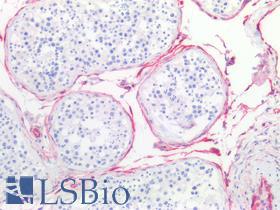
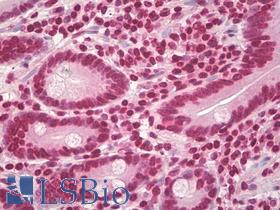
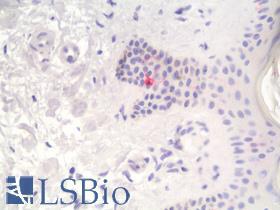
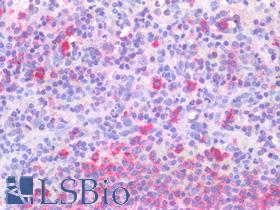
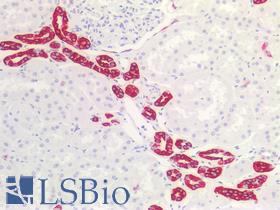
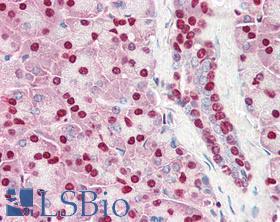
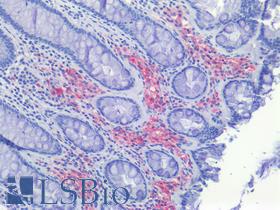
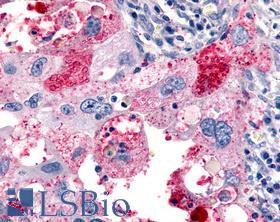
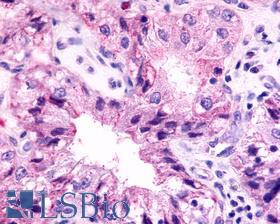
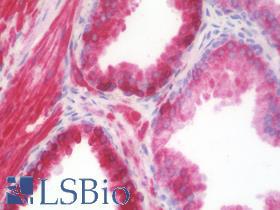
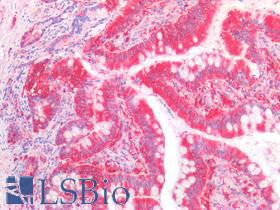
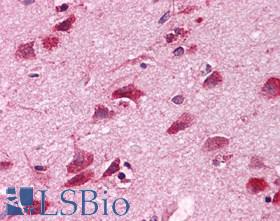
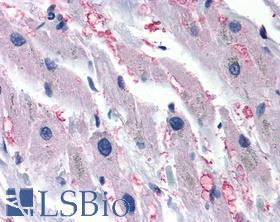
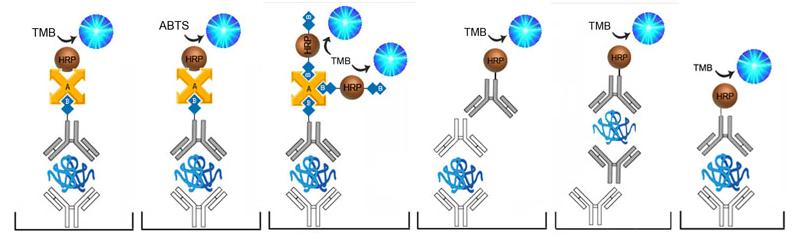
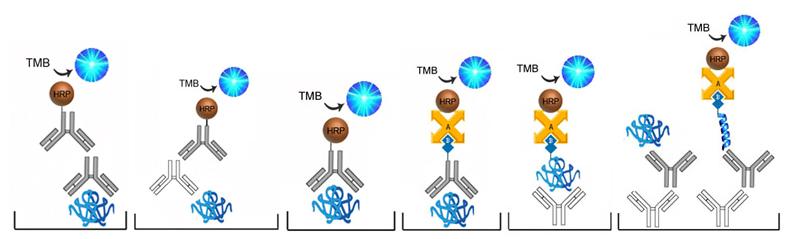
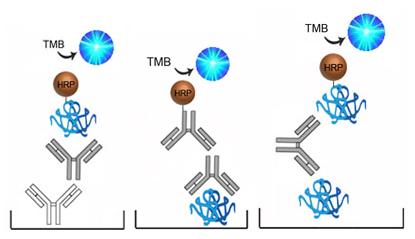
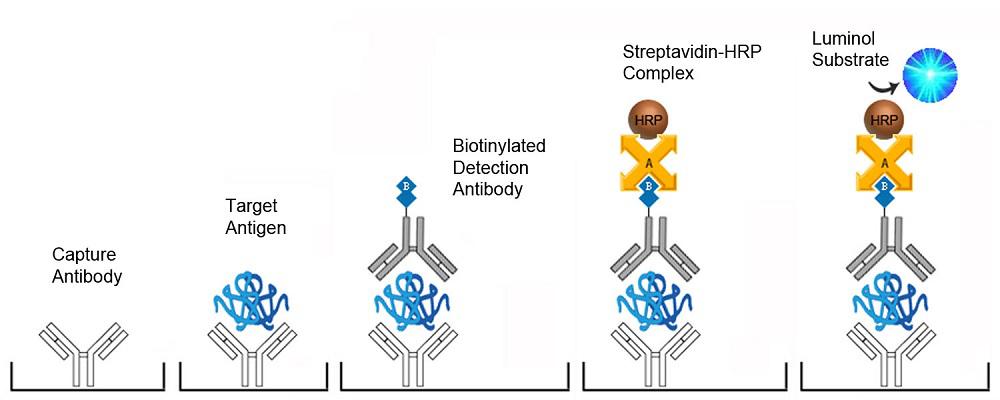
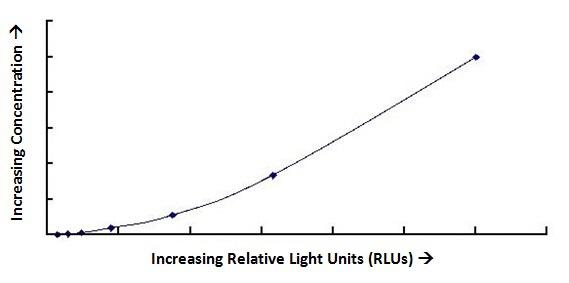
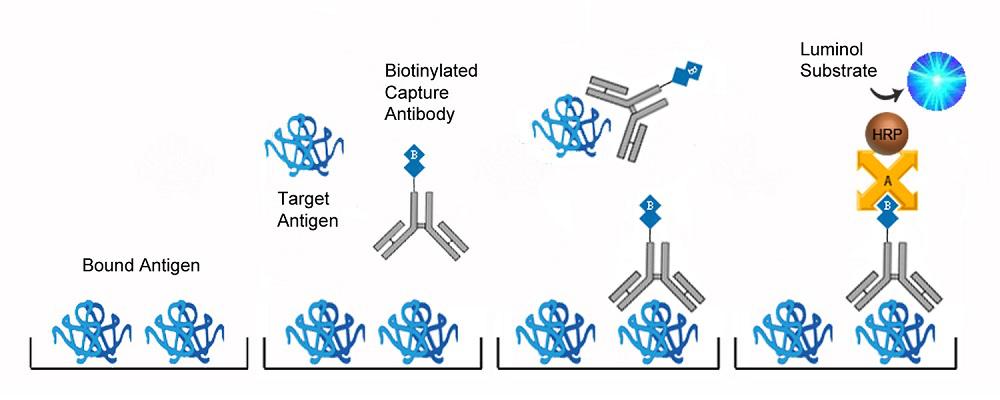
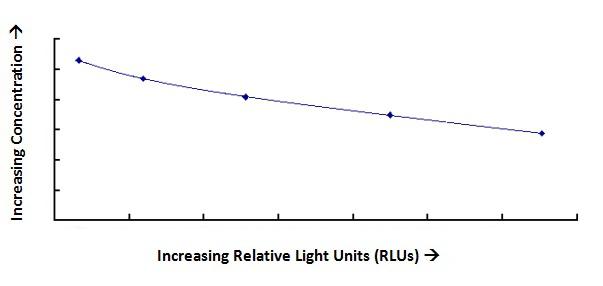
| Product Name |
Catalogue Number |
| VECTASTAIN® Elite® ABC-HRP Reagent, R.T.U. (Peroxidase, Ready-to-Use) |
LS-J1026 |
| VECTASTAIN® Elite® ABC-HRP Kit (Peroxidase, Universal), R.T.U. (Ready-to-Use) |
LS-J1027 |
| Antigen Unmasking Solution, Citric Acid Based |
LS-J1040 |
| VECTASHIELD Antifade Mounting Medium |
LS-J1032 |
| VECTASTAIN® ABC-AP Staining Kit (Alkaline Phosphatase, Rabbit IgG) |
LS-J1001 |
| VECTASTAIN® Elite® ABC-HRP Kit (Peroxidase, Rabbit IgG) |
LS-J1019 |
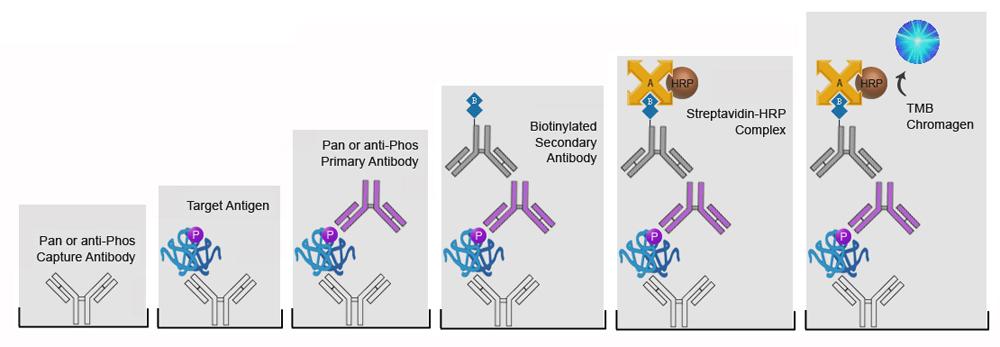
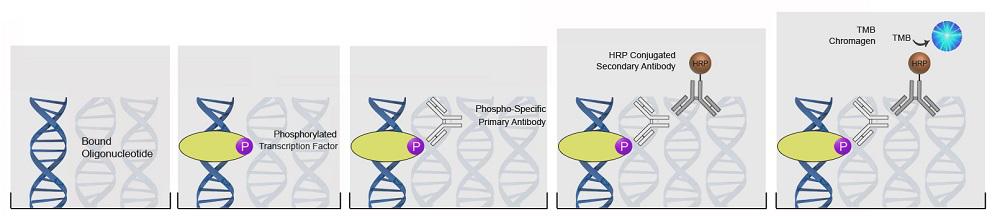
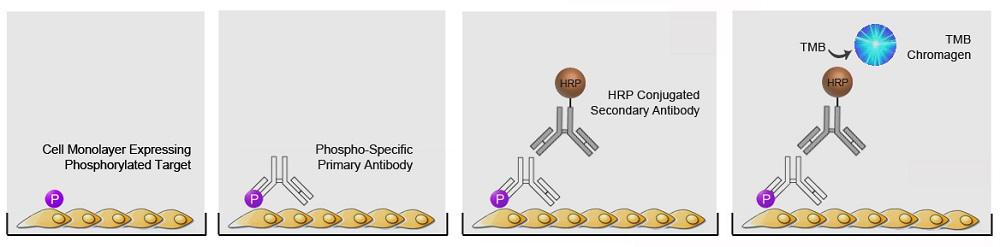
| ImmPRESS® HRP Anti-Mouse IgG (Peroxidase) Polymer Detection Kit, made in Goat |
LS-J1067 |
| ImmPACT™ DAB Peroxidase (HRP) Substrate |
LS-J1075 |
| VECTASHIELD HardSet Antifade Mounting Medium with DAPI |
LS-J1035 |
| Vector® Hematoxylin QS |
LS-J1045 |
| BLOXALL™ Endogenous Peroxidase and Alkaline Phosphatase Blocking Solution |
LS-J1031 |
| VECTASTAIN® ABC-HRP Kit (Peroxidase, Rabbit IgG) |
LS-J1010 |
| VECTASTAIN® Elite® ABC-HRP Kit (Peroxidase, Standard) |
LS-J1018 |
| VECTASTAIN® ABC-HRP Kit (Peroxidase, Standard) |
LS-J1009 |
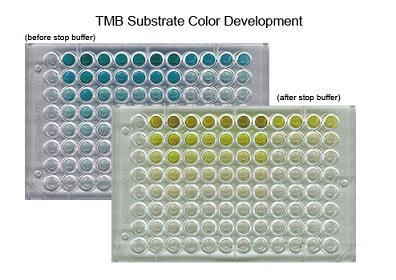
| TMB SUBSTRATE KIT |
LS-J1079 |
| Vector Red™ Alkaline Phosphatase (Red AP) Substrate Kit |
LS-J1086 |
| Hematoxylin and Eosin Stain Kit |
LS-J1047 |
| VECTASTAIN® Elite® ABC-HRP Kit (Peroxidase, Goat IgG) |
LS-J1023 |
| ImmPRESS® HRP Universal Antibody (Anti-Mouse IgG/Anti-Rabbit IgG, Peroxidase) Polymer Detection Kit, made in Horse |
LS-J1068 |
| ImmPRESS® Excel Amplified HRP Polymer Staining Kit (Anti-Rabbit IgG) |
LS-J1069 |
| ImmPRESS*-VR HRP ANTI-RABBIT IgG KIT (15 ml) |
LS-J1058 |
| ImmPRESS® HRP Anti-Rabbit IgG (Peroxidase) Polymer Detection Kit, made in Goat |
LS-J1066 |
| Vector® Nuclear Fast Red |
LS-J1044 |
| Antigen Unmasking Solution, Tris-Based |
LS-J1041 |
| Vector® Methyl Green |
LS-J1043 |
| VECTASHIELD Antifade Mounting Medium with DAPI |
LS-J1033 |
| VECTASTAIN® ABC-AP Staining Kit (Alkaline Phosphatase, Standard) |
LS-J1000 |
| VECTASTAIN® ABC-AP Staining Kit (Alkaline Phosphatase, Rat IgG) |
LS-J1004 |
| VECTASTAIN® Elite® ABC-HRP Kit (Peroxidase, Mouse IgG) |
LS-J1020 |
| VECTASTAIN® ABC-AP Staining Kit (Alkaline Phosphatase, Human IgG) |
LS-J1003 |
| VECTASTAIN® ABC-AP Staining Kit (Alkaline Phosphatase, Goat IgG) |
LS-J1005 |
| VECTASTAIN® ABC-HRP Kit (Peroxidase, Human IgG) |
LS-J1012 |
| VECTASTAIN® ABC-HRP Kit (Peroxidase, Sheep IgG) |
LS-J1015 |
| VECTASTAIN® ABC-AP Staining Kit (Alkaline Phosphatase, Mouse IgM) |
LS-J1007 |
| ImmPRESS®-AP Anti-Rabbit IgG (Alkaline Phosphatase) Polymer Detection Kit |
LS-J1053 |
| ImmPRESS®-AP Anti-Goat IgG (Alkaline Phosphatase) Polymer Detection Kit |
LS-J1056 |
| ImmPRESS®-AP Anti-Mouse IgG (Alkaline Phosphatase) Polymer Detection Kit |
LS-J1054 |
| ImmPRESS® HRP Anti-Rabbit IgG (Peroxidase) Polymer Detection Kit, made in Horse |
LS-J1060 |
| ImmPRESS® HRP Anti-Mouse IgG (Peroxidase) Polymer Detection Kit, made in Horse |
LS-J1061 |
| ImmPRESS® HRP Anti-Goat IgG (Peroxidase) Polymer Detection Kit, made in Horse |
LS-J1063 |
| ImmPRESS® Excel Amplified HRP Polymer Staining Kit (Anti-Mouse IgG) |
LS-J1070 |
| ImmPRESS® Duet Double Staining HRP/AP Polymer Kit (anti-rabbit IgG-brown, anti-mouse IgG-red) |
LS-J1071 |
| AEC Peroxidase (HRP) Substrate Kit, 3-amino-9-ethylcarbazole |
LS-J1076 |
| DAB Peroxidase (HRP) Substrate Kit (with Nickel), 3,3’-diaminobenzidine |
LS-J1073 |
| ImmPACT™ AEC Peroxidase (HRP) Substrate |
LS-J1077 |
| ImmPACT™ AMEC Red Peroxidase (HRP) Substrate |
LS-J1078 |
| ImmPRESS®-AP Anti-Rat IgG (Alkaline Phosphatase) Polymer Detection Kit, Made in Goat |
LS-J1055 |
| ImmPRESS®-AP Anti-Rat IgG, Mouse Adsorbed (Alkaline Phosphatase) Polymer Detection Kit, Made in Goat |
LS-J1057 |
| ImmPRESS*-VR HRP ANTI-MOUSE IgG KIT (15 ml) |
LS-J1059 |
| ImmPRESS® Duet Double Staining HRP/AP Polymer Kit (anti-mouse IgG-brown, anti-rabbit IgG-red) |
LS-J1072 |
| ImmPACT™ DAB EqV Peroxidase (HRP) Substrate |
LS-J1074 |
| ImmPRESS® HRP Anti-Rat IgG (Peroxidase) Polymer Detection Kit, made in Goat |
LS-J1062 |
| ImmPRESS® HRP Anti-Mouse IgG, Rat adsorbed (Peroxidase) Polymer Detection Kit, made in Horse |
LS-J1064 |
| ImmPRESS® HRP Anti-Rat IgG, Mouse adsorbed (Peroxidase) Polymer Detection Kit, made in Goat |
LS-J1065 |
| ImmPrint Permanent Marking Pen |
LS-J1049 |
| ImmPACT™ VIP Peroxidase (HRP) Substrate |
LS-J1081 |
| Vector® VIP Peroxidase (HRP) Substrate Kit |
LS-J1080 |
| Vector® SG Peroxidase (HRP) Substrate Kit |
LS-J1082 |
| ImmPACT™ SG Peroxidase (HRP) Substrate |
LS-J1083 |
| Vector® Black Alkaline Phosphatase (AP) Substrate Kit |
LS-J1088 |
| Animal-Free Blocker™ (5x) |
LS-J1090 |
| ImmPACT™ Vector Red™ Alkaline Phosphatase (AP) Substrate |
LS-J1087 |
| Vector® Blue Alkaline Phosphatase (Blue AP) Substrate Kit |
LS-J1089 |
| Vector® NovaRED™ Peroxidase (HRP) Substrate Kit |
LS-J1084 |
| ImmPACT™ NovaRED™ Peroxidase (HRP) Substrate |
LS-J1085 |
| VectaMount™ AQ Aqueous Mounting Medium |
LS-J1038 |
| DAB Enhancing Solution |
LS-J1039 |
| Alcian Blue (pH 2.5) Stain Kit |
LS-J1046 |
| Vector® Hematoxylin |
LS-J1042 |
| VECTASHIELD HardSet Antifade Mounting Medium |
LS-J1034 |
| VECTASHIELD HardSet Antifade Mounting Medium with Phalloidin |
LS-J1036 |
| VectaMount™ Permanent Mounting Medium |
LS-J1037 |
| Levamisole Solution |
LS-J1030 |
| Normal Chicken Serum Blocking Solution |
LS-M7 |
| VECTASTAIN® ABC-HRP Kit (Peroxidase, Mouse IgG) |
LS-J1011 |
| VECTASTAIN® ABC-HRP Kit (Peroxidase, Rat IgG) |
LS-J1013 |
| VECTASTAIN® ABC-HRP Kit (Peroxidase, Goat IgG) |
LS-J1014 |
| VECTASTAIN® ABC-HRP Kit (Peroxidase, Mouse IgM) |
LS-J1017 |
| VECTASTAIN® ABC-HRP Kit (Peroxidase, Guinea Pig IgG) |
LS-J1016 |
| Normal Goat Serum Blocking Solution |
LS-M1 |
| 2.5% Normal Goat Serum Blocking Solution |
LS-M2 |
| Normal Horse Serum Blocking Solution |
LS-M3 |
| VECTASTAIN® ABC-AP Staining Kit (Alkaline Phosphatase, Mouse IgG) |
LS-J1002 |
| 2.5% Normal Horse Serum Blocking Solution |
LS-M4 |
| VECTASTAIN® Elite® ABC-HRP Kit (Peroxidase, Human IgG) |
LS-J1021 |
| VECTASTAIN® Elite® ABC-HRP Kit (Peroxidase, Rat IgG) |
LS-J1022 |
| VECTASTAIN® Elite® ABC-HRP Kit (Peroxidase, Sheep IgG) |
LS-J1024 |
| VECTASTAIN® Elite® ABC-HRP Kit (Peroxidase, Universal) |
LS-J1025 |
| Normal Swine Serum Blocking Solution |
LS-M5 |
| VECTASTAIN® ABC-AP Staining Kit (Alkaline Phosphatase, Universal) |
LS-J1008 |
| Normal Rabbit Serum Blocking Solution |
LS-M6 |