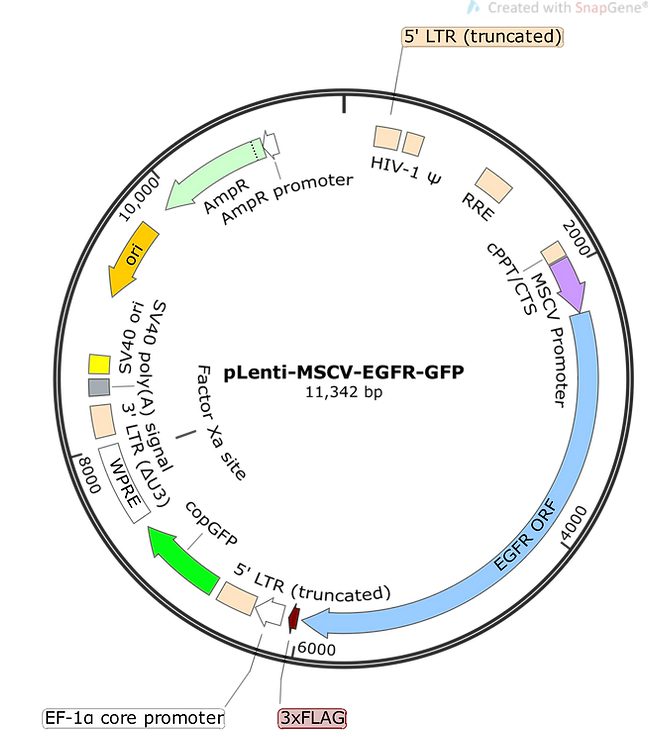
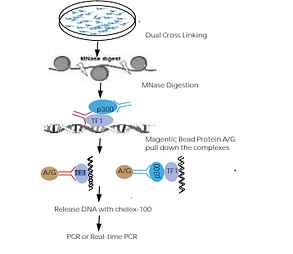
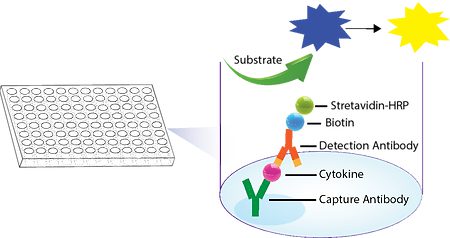
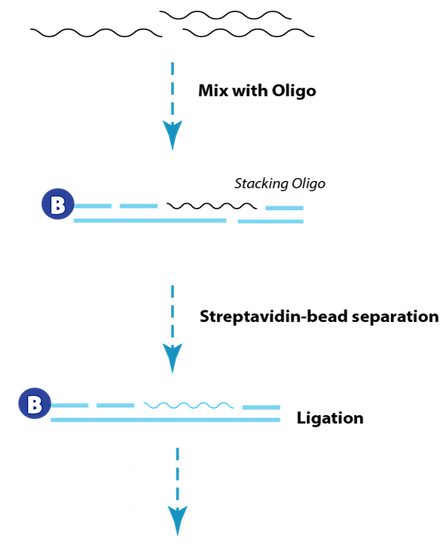
| Description |
Catalog No. |
| NFkB Secreted Luciferase Reporter HeLa Stable Cell Line |
SL-4001 |
| NFkB Secreted Luciferase Reporter HEK293 Stable Cell Line |
SL-4012 |
| NFkB Luciferase Reporter HepG2 Stable Cell Line |
SL-0017 |
| ELK-TAD Luciferase Reporter HEK293 Stable Cell Line |
SL-0040 |
| TCF/LEF Luciferase Reporter HEK293 Stable Cell Line |
SL-0015 |
| Stat1 Luciferase Reporter Hela Stable Cell Line |
SL-0004 |
| NFkB Luciferase Reporter HEK293 Stable Cell Line |
SL-0012 |
| NFkB Luciferase Reporter MCF7 Stable Cell Line |
SL-0013 |
| Estrogen Receptor Luciferase Reporter T47D Stable Cell Line |
SL-0002 |
| NFkB Luciferase Reporter HeLa Stable Cell Line |
SL-0001 |
| p53 Luciferase Reporter Hela Stable Cell Line |
SL-0011 |
| SMAD/TGFbeta Luciferase Reporter HepG2 Stable Cell Line |
SL-0016 |
| AR Luciferase Reporter MDA-MB-453 Stable Cell Line |
SL-0008 |
| NRF2/ARE Luciferase Reporter MCF7 Stable Cell Line |
SL-0010 |
| NFkB Luciferase Reporter NIH 3T3 Stable Cell Line |
SL-0006 |
| GR Luciferase Reporter MDA-MB-453 Stable Cell Line |
SL-0009 |
| p53 Luciferase Reporter RKO Stable Cell Line |
SL-0007 |
| HIF Luciferase Reporter NIH3T3 Stable Cell Line SL-0005 |
SL-0005 |
| NFkB Luciferase Reporter A549 Stable Cell Line |
SL-0014 |
| Control HepG2 Luciferase Reporter Stable Cell Line |
SL-0039 |
| Control Hela Luciferase Reporter Stable Cell Line |
SL-0038 |
| SMAD/BMP Responsive Luciferase Reporter HEK293 Stable Cell Line |
SL-0051 |
| CREB Luciferase Reporter HEK293 Stable Cell Line |
SL-0020 |
| Glucocorticoid Receptor Luciferase Reporter Hela Stable Cell Line |
SL-0021 |
| FXR Luciferase Reporter HepG2 Stable Cell Line |
SL-0055 |
| NFkB Luciferase Reporter MEF Stable Cell Line |
SL-0033 |
| NRF2/ARE Luciferase Reporter HepG2 Stable Cell Line |
SL-0046 |
| MRF Luciferase Reporter HEK293 Stable Cell Line |
SL-0053 |
| NFAT Luciferase Reporter Hela Stable Cell Line |
SL-0018 |
| TCF/LEF Luciferase Reporter Hela Stable Cell Line |
SL-0022 |
| HIF Luciferase Reporter Neuro2a Stable Cell Line |
SL-0027 |
| NFkB Luciferase Reporter Jurkat Stable Cell Line |
SL-0050 |
| NRF2/ARE Luciferase Reporter NIH3T3 Stable Cell Line |
SL-0047 |
| ELK-TAD Luciferase Reporter Hela Stable Cell Line |
SL-0041 |
| CHOP Luciferase Reporter Mia_Paca2 Stable Cell Line |
SL-0025 |
| NRF2/ARE Luciferase Reporter HEK293 Stable Cell Line |
SL-0042 |
| IRF Luciferase Reporter HEK293 Stable Cell Line |
SL-0035 |
| ATF6 Luciferase Reporter CHO-K1 Stable Cell Line |
SL-0024 |
| NFkB Luciferase Reporter MDA-MB-231 Stable Cell Line |
SL-0043 |
| TCF/LEF Luciferase Reporter CHO-K1 Stable Cell Line |
SL-0028 |
| NFkB Luciferase Reporter Neuro2a Stable Cell Line |
SL-0026 |
| CREB Luciferase Reporter NIH/3T3 Stable Cell Line |
SL-0031 |
| IFNa-Luciferase Reporter HeLa Stable Cell Line |
SL-0052 |
| HIF Luciferase Reporter Hela Stable Cell Line |
SL-0023 |
| SMAD/TGFbeta Luciferase Reporter NIH/3T3 Stable Cell Line |
SL-0030 |
| IRF Luciferase Reporter HepG2 Stable Cell Line |
SL-0049 |
| HIF Luciferase Reporter Cos-7 Stable Cell Line |
SL-0034 |
| NFAT Luciferase Reporter Jurkat Stable Cell Line |
SL-0032 |
| AP-1 Luciferase Reporter Hela Stable Cell Line |
SL-0019 |
| NFAT Luciferase Reporter NIH 3T3 Stable Cell Line |
SL-0029 |
| ATF4 Luciferase Reporter HEK293 Stable Cell Line |
SL-0080 |
| ATF6 Luciferase Reporter HEK293 Stable Cell Line |
SL-0084 |
| Amino acid deprivation-AARE Luciferase Reporter 293 Stable Cell Line |
SL-0066 |
| Amino acid deprivation-AARE Luciferase Reporter MEF Stable Cell Line |
SL-0065 |
| ER Stress-ERSE Luciferase Reporter 293 Stable Cell Line |
SL-0068 |
| NFAT Luciferase Reporter HEK293 Stable Cell Line |
SL-0078 |
| ELK Luciferase Reporter HEK293 Stable Cell Line |
SL-0077 |
| AP1 Luciferase Reporter HEK293 Stable Cell Line |
SL-0085 |
| XRE Luciferase Reporter HEK293 Stable Cell Line |
SL-0075 |
| Stat3 Luciferase Reporter HEK293 Stable Cell Line |
SL-0071 |
| GAS/Stat1 Luciferase Reporter HCT116 Stable Cell Line |
SL-0074 |
| ATF4 Luciferase Reporter HepG2 Stable Cell Line |
SL-0079 |
| ATF6 Luciferase Reporter HepG2 Stable Cell Line |
SL-0082 |
| ER Stress Luciferase Reporter HepG2 Stable Cell Line |
SL-0069 |
| Stat3 Luciferase Reporter HepG2 Stable Cell Line |
SL-0081 |
| ER Stress-ERSE Luciferase Reporter MEF Stable Cell Line |
SL-0067 |
| CHOP Luciferase Reporter HEK293 Stable Cell Line |
SL-0083 |
| p53 Luciferase Reporter HCT116 Stable Cell Line |
SL-0072 |