| Protein | Functions | References |
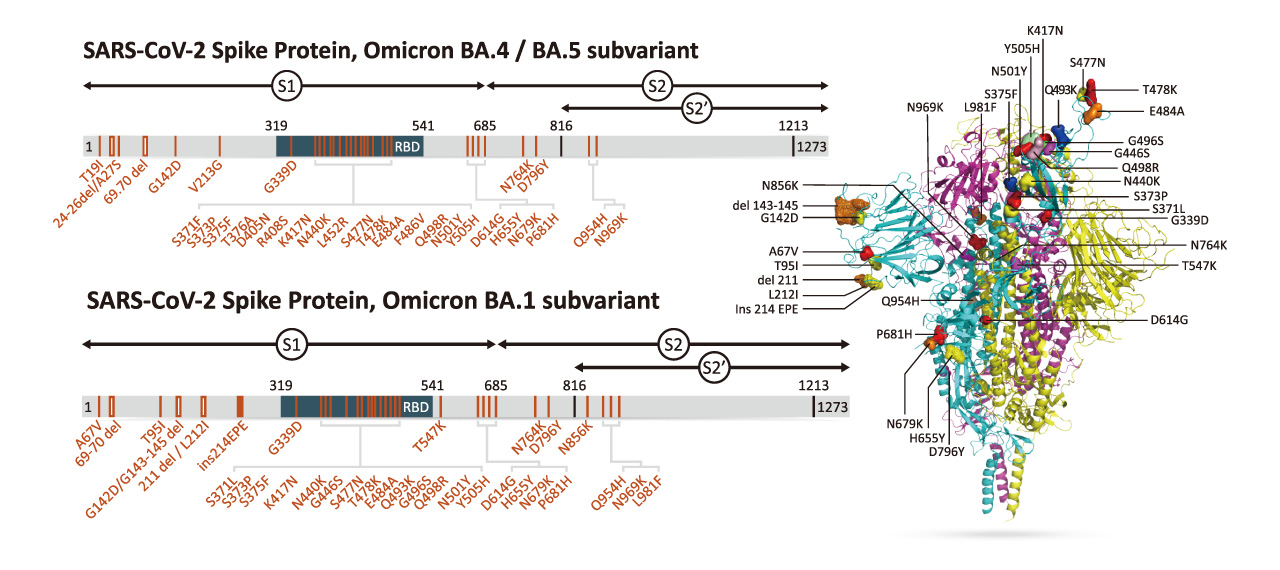
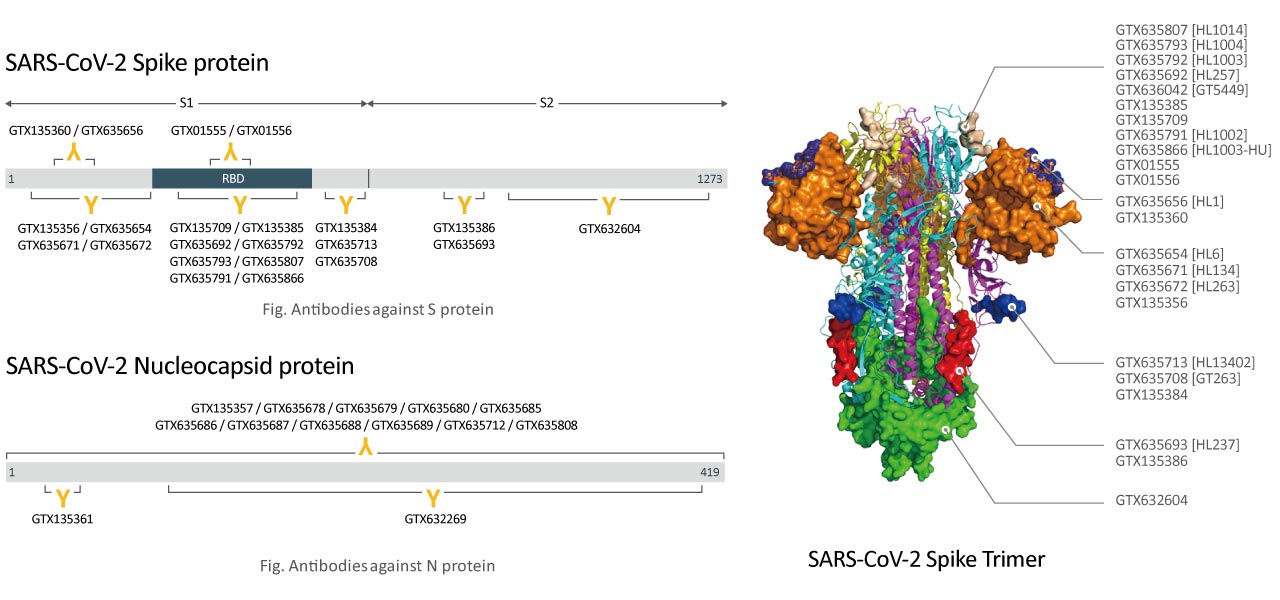
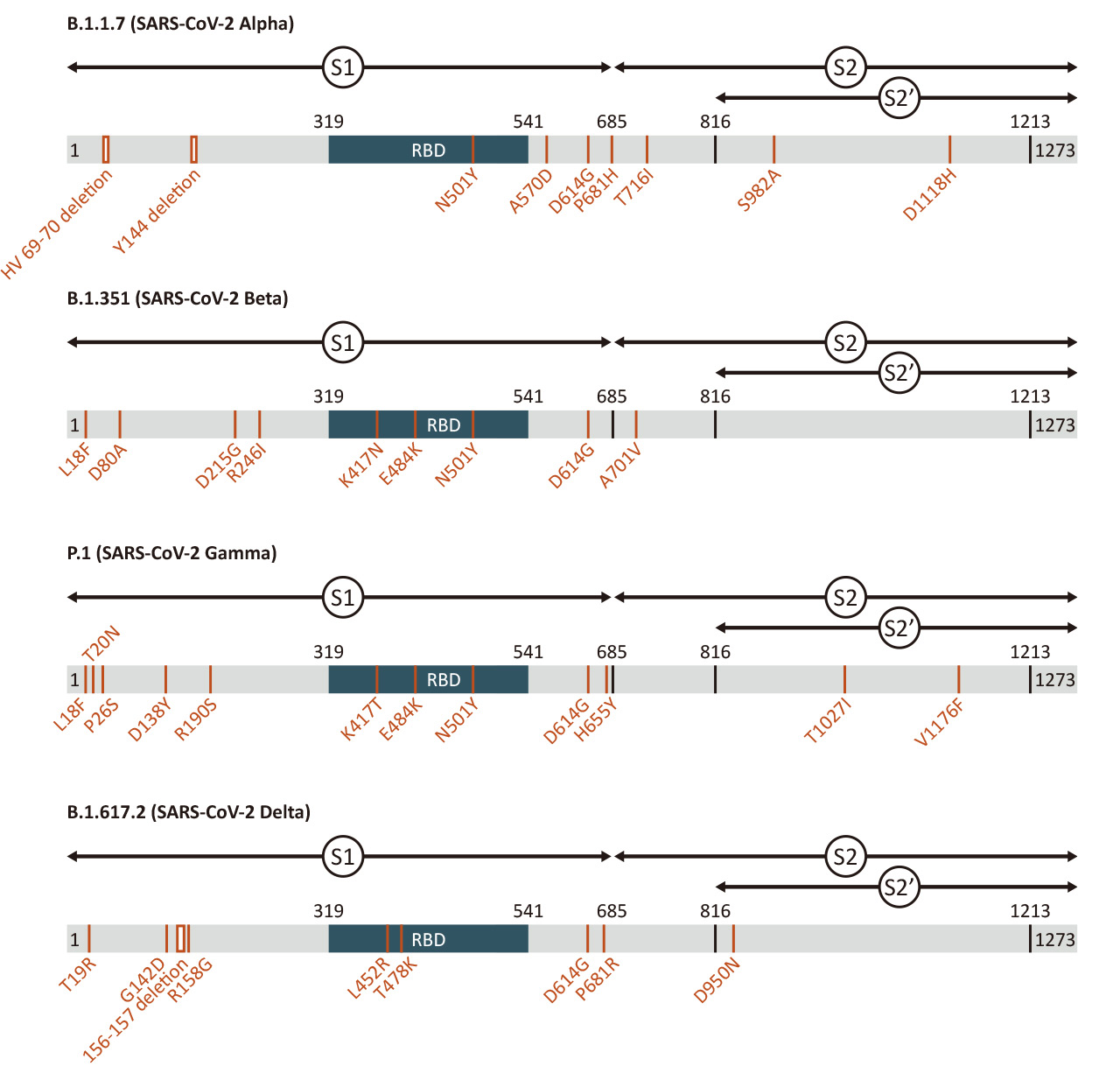
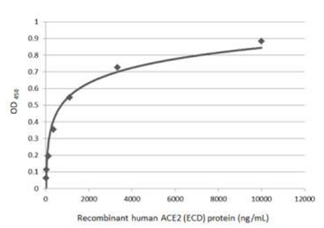
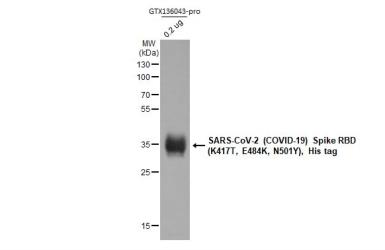
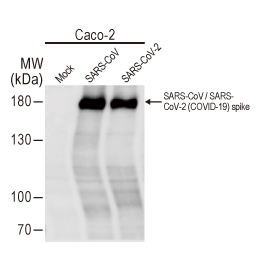
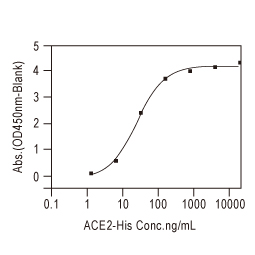
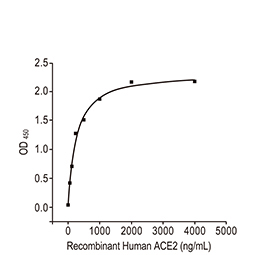
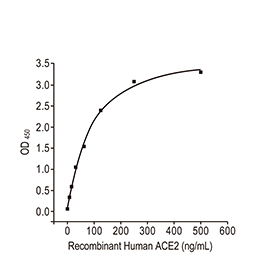
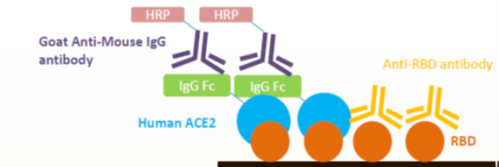
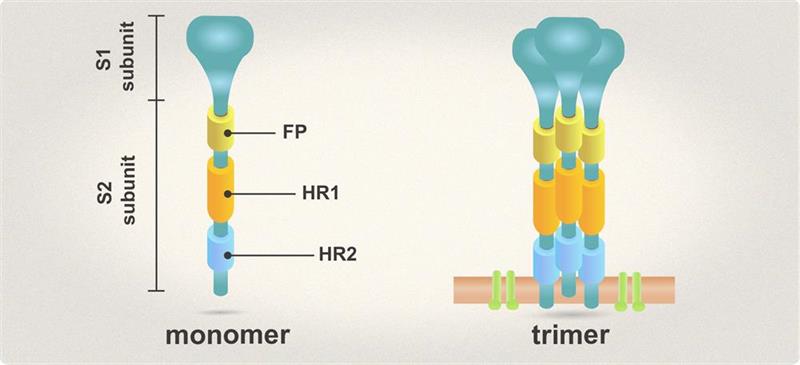
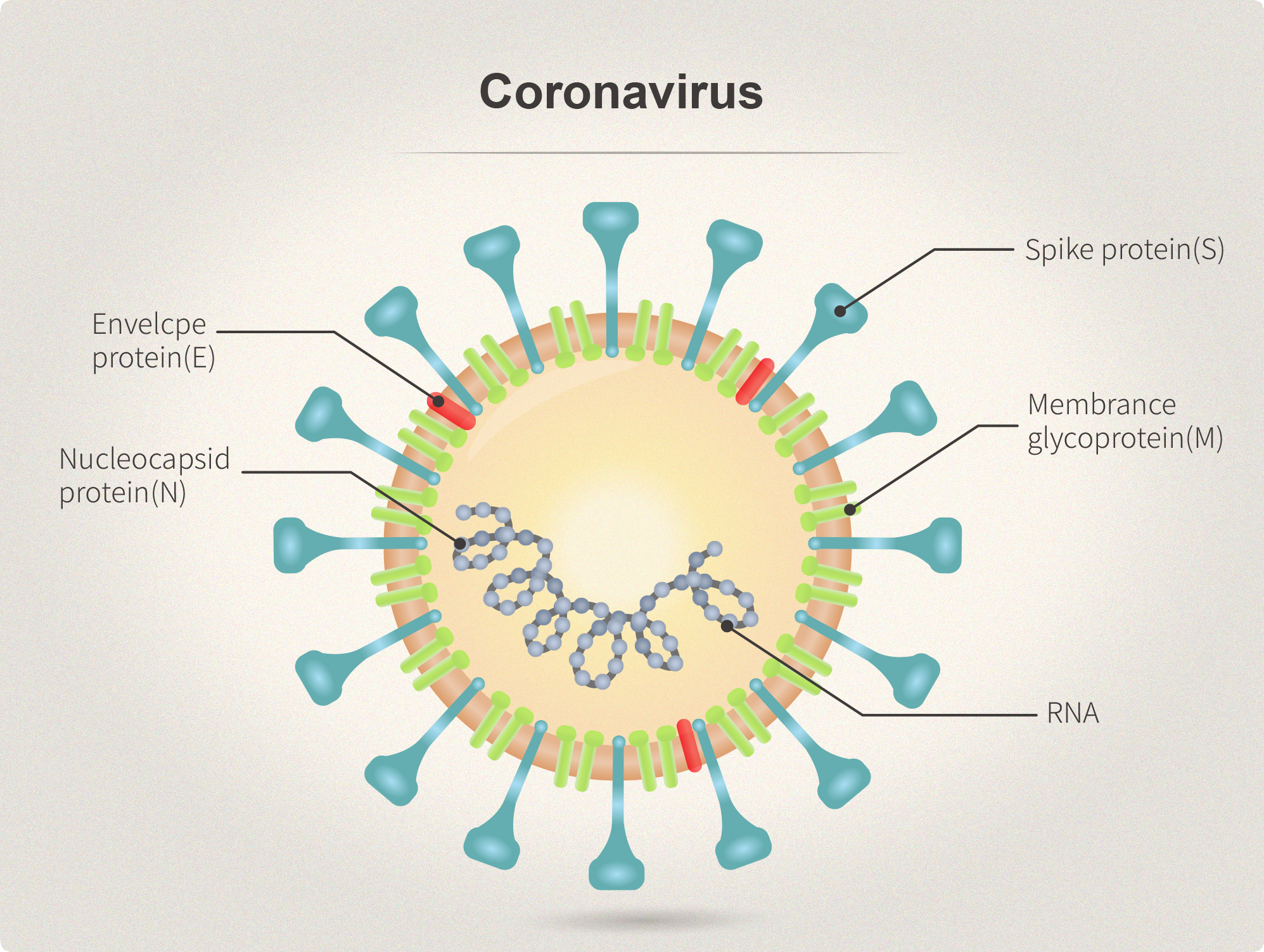
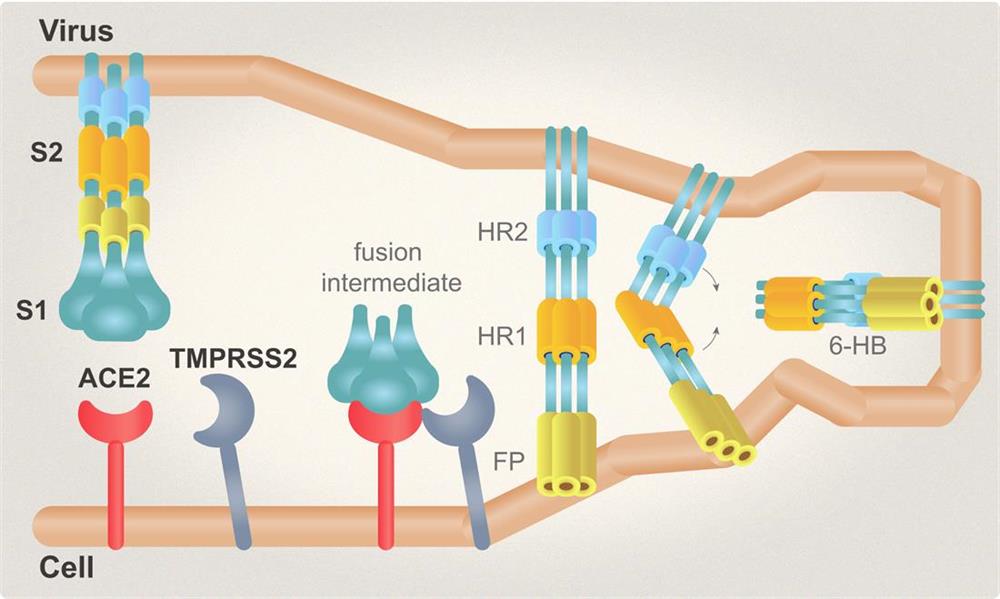
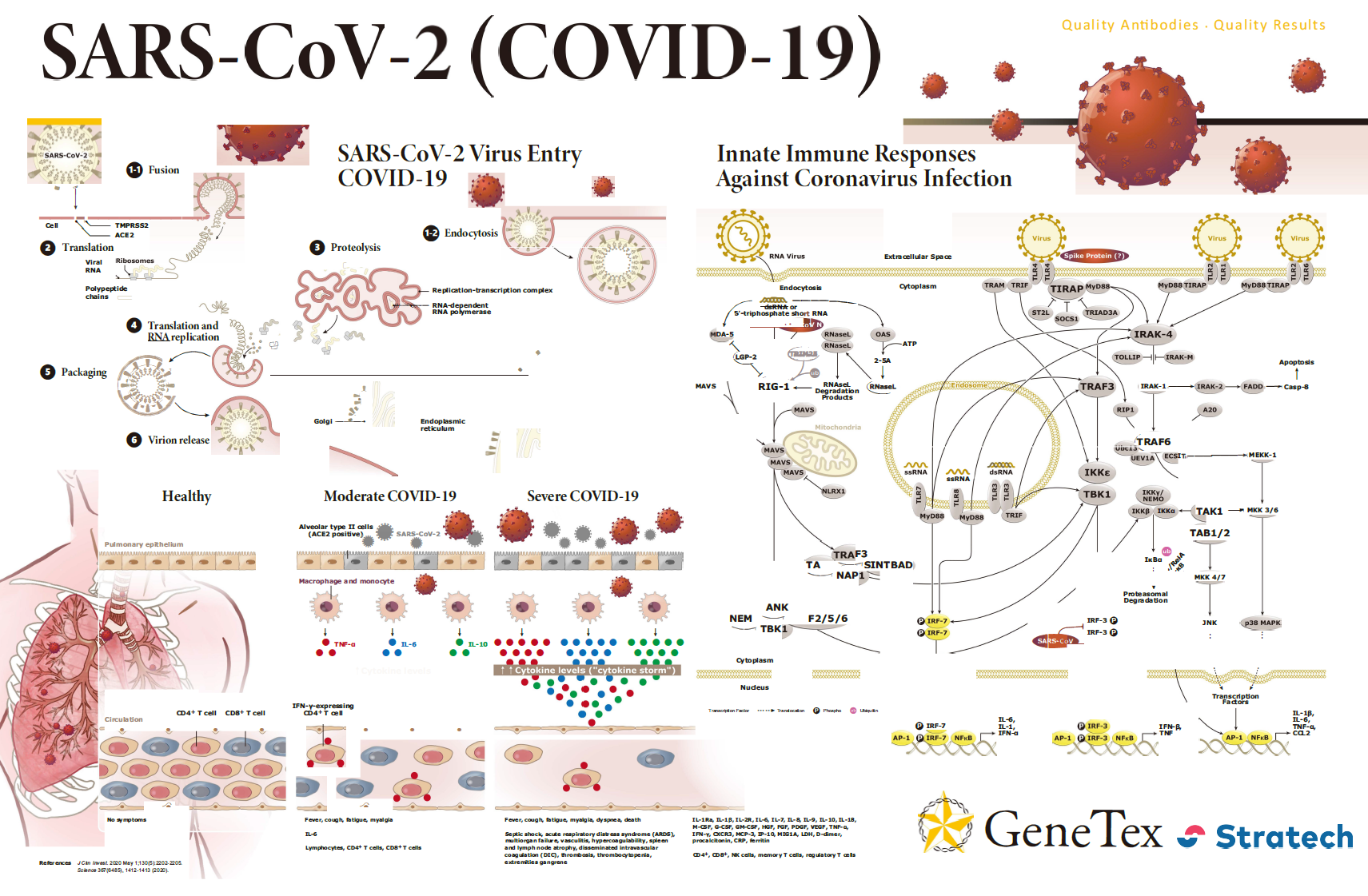
| Spike (S) | Spike full-length (~1273 a.a. in SARS-CoV-2) protein precursor is cleaved into glycosylated subunits, S1 and S2 (S2’). S1 binds to the host’s receptor, ACE2, while S2 mediates viral and host membrane fusion. | 1 |
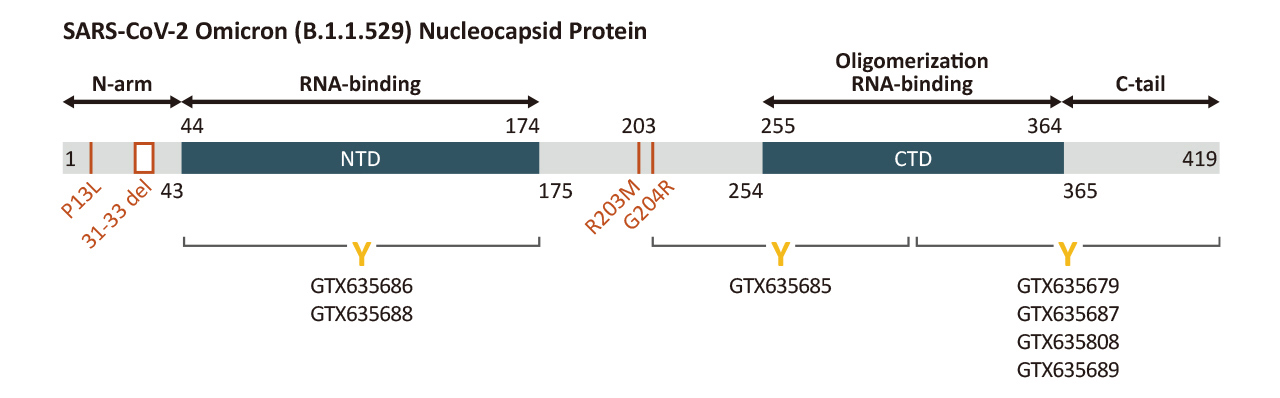
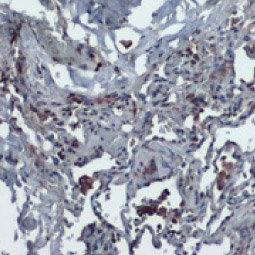
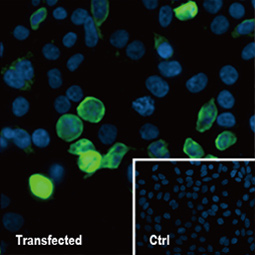
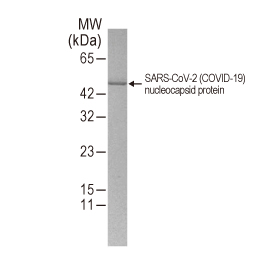
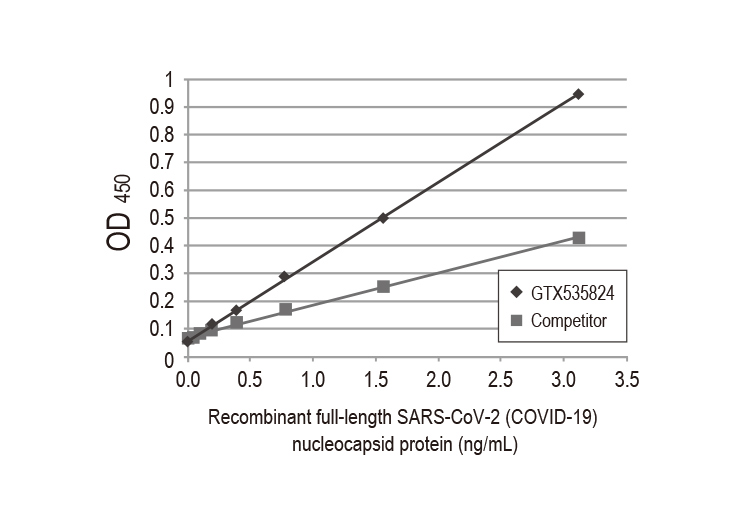
| Nucleocapsid (N) | Nucleocapsid (~419 a.a. in SARS-CoV-2) binds viral genomic RNA and forms a helical ribonucleocapsid. Involved in genome protection, viral RNA replication, virion assembly, and immune evasion. Interacts with M and nsp3 proteins. | 2 |
| Membrane (M) | Membrane/matrix protein (~222 a.a. in SARS-CoV-2) is the most abundant structural component of the virion, and very conserved. Mediates assembly and budding of viral particles through recruitment of other structural proteins to “ER-Golgi-intermediate compartment (ERGIC)”. Interaction with N for RNA packaging into virion. Interacts with accessory proteins 3a and 7a. Mitigation of immune response? | 3 |
| Envelope (E) | Envelope small membrane protein (~75 a.a. in SARS-CoV-2) is a single-pass type III membrane protein involved in viral assembly, budding, and pathogenesis. Localizes to ERGIC. Forms a homopentameric ion channel and is a viroporin. Interacts with M, N, 3a, and 7a. | 4 |
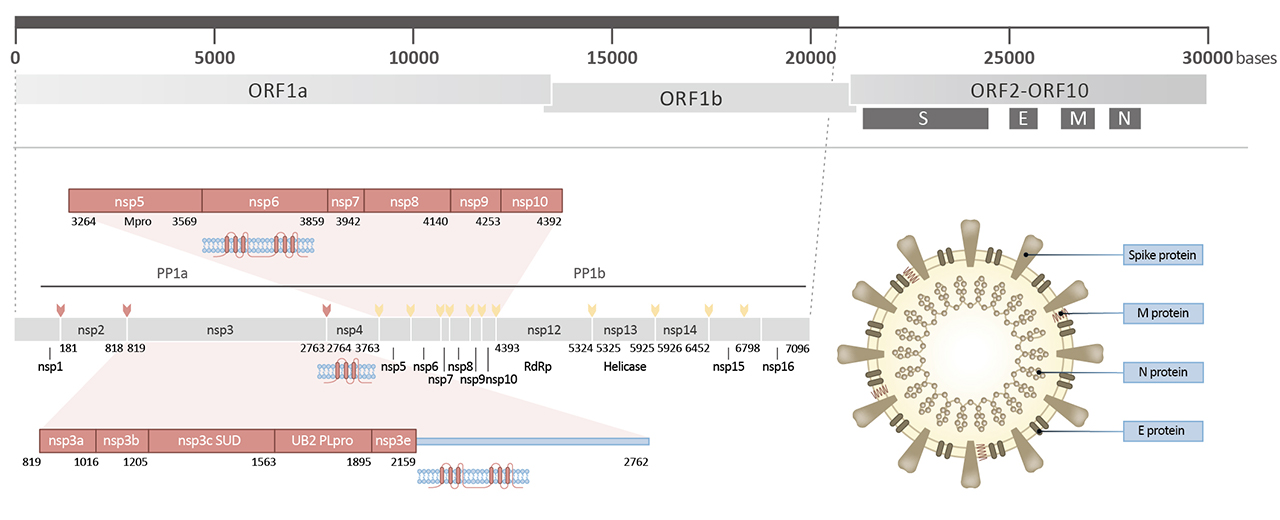
| nsp1 | Nonstructural protein 1 (nsp1; ~180 a.a. in SARS-CoV-2) likely inhibits host translation by interacting with 40S ribosomal subunit, leading to host mRNA degradation through cleavage near their 5’UTRs. Promotes viral gene expression and immunoevasion in part by interfering with interferon-mediated signaling. | 5 |
| nsp2 | nsp2 (~638 a.a. in SARS-CoV-2) interacts with host factors prohibitin 1 and prohibitin 2, which are involved in many cellular processes including mitochondrial biogenesis. It appears that nsp2 may change the intracellular milieu and perturb host intracellular signaling. | 6 |
| nsp3 | nsp3 (~1945 a.a. in SARS-CoV-2) is a papain-like protease (PLpro) and multi-pass membrane protein that processes the viral polyprotein to release nsp1, nsp2, and nsp3. It also exhibits deubiquitinating and deISGylating activities. Interacts with nsp4 and nsp6. | 7 |
| nsp4 | nsp4 (~500 a.a. in SARS-CoV-2) is required for viral replication by inducing (with nsp3) assembly of, and localizing to, double-membrane cytoplasmic vesicles. Multi-pass membrane protein. | 8 |
| nsp5 | nsp5 (3CLpro; ~306 a.a. in SARS-CoV-2) cleaves at 11 sites in the polyprotein to release nsp4-nsp16. It is also responsible for nsp maturation. | 9 |
| nsp6 | nsp6 (~290 a.a. in SARS-CoV-2) is a multi-pass membrane protein that induces double-membrane vesicles in infected cells with nsp 3 and nsp4. It also limits autophagosome expansion and interferes with autophagosome delivery of viral factors to lysosomes for destruction. | 10,11 |
| nsp7 | nsp7 (~83 a.a. in SARS-CoV-2) forms a hexadecamer with nsp8 as a cofactor for the RNA-dependent RNA polymerase nsp12. May have processivity or RNA primase function. | 12 |
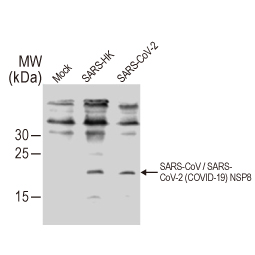
| nsp8 | nsp8 (~198 a.a. in SARS-CoV-2) forms a hexadecamer with nsp7 as a cofactor for the RNA-dependent RNA polymerase nsp12. May have processivity or RNA primase function. Mutation of certain residues in nsp8 is lethal to SARS-CoV by impacting RNA synthesis. | 13 |
| nsp9 | nsp9 (~113 a.a. in SARS-CoV-2) functions in viral replication as a dimeric ssRNA-binding protein. | 13 |
| nsp10 | nsp10 (~139 a.a. in SARS-CoV-2) forms a dodecamer and interacts with both nsp14 and nsp16 to stimulate their respective 3’-5’ exoribonuclease and 2’-O-methyltransferase activities in the formation of the viral mRNA capping machinery. | 13 |
| nsp11 | nsp11 (~13-23 a.a., depending on the CoV species) is a pp1a cleavage product at the nsp10/11 boundary. For pp1ab, it is a frameshift product that becomes the N-terminal of nsp12. Its function, if any, is unknown. | 13 |
| nsp12 | nsp12 (~932 a.a. in SARS-CoV-2) is the RNA-dependent RNA polymerase (RdRp) performing both replication and transcription of the viral genome. It has >95% identity to the SARS-CoV polymerase and is inhibited by the nucleoside analogue Remdesivir. | 13 |
| nsp13 | nsp13 (~601 a.a. in SARS-CoV-2) is a multifunctional superfamily 1 helicase capable of using both dsDNA and dsRNA as substrates with 5’-3’ polarity. In addition to working with nsp12 in viral genome replication, it is also involved in viral mRNA capping. It associates with nucleoprotein in membranous complexes. | 14 |
| nsp14 | nsp14 (~527 a.a. in SARS-CoV-2) has both 3’-5’ exoribonuclease (proofreading during RNA replication) and N7-guanine methyltransferase (viral mRNA capping) activities. Interacts with nsp10. | 13 |
| nsp15 | nsp15 (~346 a.a. in SARS-CoV-2) is an endoribonuclease that favors cleavage of RNA at the 3’-ends of uridylates. Loss of nsp15 affects both viral replication and pathogenesis. It is also required for evasion of host cell dsRNA sensors. | 15 |
| nsp16 | nsp16 (~298 a.a. in SARS-CoV-2) interacts with and is activated by nsp10. Its 2’-O-methyltransferase activity is essential for viral mRNA capping. It may also work against host cell antiviral sensors. | 13 |
| ORF3a | ORF3a (~275 a.a. in SARS-CoV-2) is a multi-pass membrane protein that forms a homotetrameric viroporin in SARS-CoV. It interacts with accessory protein 7a, M, S and E. May be involved in viral release. Importantly, it also activates both NF-kB and NLRP3 inflammasome and contributes to the generation of cytokine storm. | 16 |
| ORF6 | ORF6 (~61 a.a. in SARS-CoV-2) appears to be a virulence factor in SARS-CoV. It was shown to be an antagonist of type I interferons (IFNs) and is involved in viral escape from the host innate immune system. | 17 |
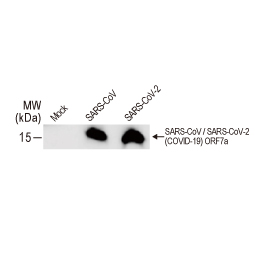
| ORF7a | ORF7a (~121 a.a. in SARS-CoV-2) is a type I membrane protein that interacts with bone marrow stromal antigen 2 (BST-2) in SARS-CoV. BST-2 tethers virions to the host’s plasma membrane. ORF7a binding inhibits BST-2 glycosylation and interferes with this restriction activity. ORF7a also interacts with S, M, E, and ORF3a in SARS-CoV. | 18 |
| ORF7b | ORF7b (~43 a.a. in SARS-CoV-2) is a type III integral transmembrane protein in the Golgi apparatus. In SARS-CoV, it appears to be a viral attenuation factor. | 19 |
| ORF8 | ORF8 (~121 a.a. in SARS-CoV-2) has only 30% identity to the intact ORF8 of SARS-CoV and might be a luminal ER membrane-associated protein. It may trigger ATF6 activation and affect the unfolded protein response (UPR). | 20,21,22 |
| ORF9b | ORF9b (~97 a.a. in SARS-CoV-2) is coded for in an alternative ORF within the N gene. No function is known, though the SARS-CoV protein interacts with nsp5, nsp14, and ORF6. There is limited evidence it may bind to lipids. | 23 |
| ORF10 | ORF10 (~38 a.a. in SARS-CoV-2) has no known function but might have a regulatory role involving interaction with another factor(s). | 24 |
| Protein | Functions | References |